
Distinct metabolic changes in human lung cancer cells with differential radiation sensitivities
Introduction
Lung cancer is the leading cause of cancer death in the United States and many other countries in the world. It accounted for 26% and 30% of all female and male cancer-related deaths, respectively, and a total of 160,000 deaths in the United States in 2009 (1). The majority of lung tumors are non-small cell lung carcinomas (NSCLCs), particularly adenocarcinoma and squamous cell carcinoma (2). Radiotherapy is one of the most important modalities for cancer treatment (3-5), particularly for elderly patients, because they generally have multiple co-morbidities and/or poor performance status (6,7), thus excluding surgery and/or chemo-radiation therapy as treatment options. However, the induction of radio-resistance and the subsequent proliferation of tumor cells have become the major causes of radiation treatment failure. Although DNA repair signaling plays an important role in inducing radio-resistance (8), more molecular mechanisms that contribute to radio-resistance need to be clarified.
Compared to genomics and proteomics, metabolomics is an emerging science. It represents the phenotype of the cellular responses to pathophysiological stimuli or genetic modification (9). The metabolite profile comprises hundreds to thousands of endogenous organic metabolites. Changes in the metabolite profile can be used to understand the perturbation of a biochemical pathway in an organism. Several biochemical pathways have been found to be disturbed in response to radiation, including energy utilization (10), detoxification (11,12), osmoregulation (13), and synthesis and degradation of cellular membranes (14). However, metabolite profiles related to radio-resistance have not been well addressed.
Two human lung cancer cell lines were investigated in the present study: CL1-5 and CL1-0. The CL1-5 cell line was derived from the human lung adenocarcinoma CL1-0 cell line using a transwell invasion chamber to select progressively more invasive cancer cell populations. The invasive ability of the CL1-5 cell line is approximately six-fold higher than that of CL1-0 (15). Analysis of differential gene expression patterns indicated that the expression levels of invasion-related genes, such as proteases, adhesion molecules, cytoskeletal proteins, motility proteins, cell cycle regulators, and signal transduction molecules, differed between the CL1-0 and CL1-5 cell lines (16). Moreover, the previous study indicated that CL1-5 cells are more sensitive than the CL1-0 cells in their response to radiation (17). Therefore, the two cell lines provide a cell model to further investigate metabolite profile changes depending on different radiation exposure levels.
Based on differential radiation sensitivity between the two cell lines, we hypothesized that these cells may exhibit different metabolic profiles both before and after radiation exposure. Therefore, we used proton nuclear magnetic resonance (1H-NMR) to analyze the metabolic profiles of the CL1-0 and CL1-5 cell lines in the study. We further investigated the responses of these two cell lines following exposure to radiation. The possible mechanisms of radio-resistance are discussed, which may provide new insights for radiotherapy.
Methods
Cell culture and radiation treatment
Two human lung cancer cell lines, CL1-0 and CL1-5, were maintained in RPMI1640 medium supplemented with 10% fetal bovine serum (Gibco BRL, Gaithersburg, MD, USA), 100 units/mL penicillin (Invitrogen, Carlsbad, CA, USA), and 100 g/mL streptomycin in a humidified atmosphere containing 5% CO2 at 37 °C. Cells were plated into 100 mm dishes, incubated overnight, and then treated with single doses of 10-Gy radiation using a cobalt 60 (Picker V9) source at dose rates between 200 and 250 cGy/min. Cells were harvested at 0, 1, 4, and 24 hr after irradiation for further study.
Colony formation assay
Following radiation treatment, different numbers of lung cancer cells treated or untreated with exposure to irradiation at 2, 5, or 10 Gy were seeded into 10 cm dishes for the colony formation assay. Colonies formed after 12 days of incubation were counted. Cells were first washed with PBS, fixed with methanol and acetic acid (3:1, v/v) for 1 hr, stained with crystal violet (0.1%) for 1 hr, and then washed with water. The surviving cell fraction was determined by dividing the plating efficiency of the treated culture by the plating efficiency of the CL1-0 and CL1-5 cells with or without treatment.
Sample preparation for 1H-NMR spectroscopy
Cells were harvested at the indicated time points post-irradiation (n=3). The culture medium was quickly removed, and the cells were washed twice with 1 mL ice-cold PBS. After washing with PBS solution, the remaining cells were placed on ice and then suspended in 1 mL ice-cold MeOH and deionized (DI) water (1:1, v/v) for 10 min. The extract was transferred to an eppendorf tube and centrifuged at 1,800 ×g at 4 °C for 15 min. The supernatant was collected and stored at −80 °C until use. The cell extract was thawed at room temperature and evaporated to dryness using nitrogen gas. The sample was reconstituted with 200 µL D2O containing 1 mg/mL of 3-trimethylsilyl-(2,2,3,3-2H4)-1-propionate (TMSP). The D2O provided an NMR lock signal for the NMR spectrometer. Two hundred microliters of the sample was then transferred into a NMR tube (2 mm) for the analysis.
NMR analysis and data pre-processing
Conventional 1H-NMR spectra of the cell extraction samples were obtained using a Bruker Avance 600 spectrometer (Bruker Biospin, Rheinstetten, Germany) operated at 600.04 MHz at 25 °C. The one-dimensional 1H-NMR spectra were acquired using a standard NOESYPR1D pulse sequence (recycledelay-90°-t1-90°-tm-90°-acquisition; XWIN-NMR3.5) with a recycle delay time of 2 s and a mixing time of 150 ms. The 90° pulse length was adjusted to ~4 µs at 0.17 dB, and t1 was set to 4 µs, which provided an acquisition time of 2.73 s. For each sample, 128 free induction decays (FID) were collected using 32k data points within 10 ppm, and the total data collection time was 11 min. FIDs were then multiplied by an exponential weighting function corresponding to a line broadening of 0.3 Hz, and the data were zero-filled to 64k data points.
All acquired FIDs were Fourier transformed, phase corrected, and aligned to the chemical shift of the alpha-glucose anomeric doublet at 5.223 ppm using ACD/Labs v. 10.0 1D NMR Manager (Advanced Chemistry Development, Inc., Canada). The FIDs were further imported into R v. 2.8.1 for water deletion, scaling, baseline correction and normalization. The region of the peak containing H2O was removed within 4.5 to 5 ppm. Spectral intensities were scaled to the ratio of TMSP intensity at unit resolution in each NMR spectra. Then, an in-house baseline correction process and robust mean normalization were applied to each spectrum. The spectra region within 0.2 to 4.4 ppm was binned into 420 bins with a binning size of 0.01 ppm.
NMR data analysis
A principal component analysis (PCA) of the spectral binning data was performed using R v. 2.8.1. PCA is an unsupervised method of analysis and serves to project the data set to a new set of orthogonal variables known as principal components (PCs). These PCs are related to the original data set because each PC is caudated by a linear combination (loading) of the original variables (18). It has frequently been applied to summarize the similarities and differences between multiple NMR spectra (19-21). The PCA score plots demonstrate the clustering time-dependence of CL1-0 and CL1-5 cells, and the PCA loadings indicate potential statistically significant peak regions. Metabolite identification was performed according to the PCA loadings of PC1 and PC2.
Results
Determination of cell survival rate
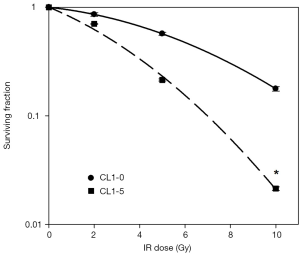
Previous studies have indicated that CL1-5 cells are more sensitive than CL1-0 cells to irradiation (17,22). Our colony formation assay results show that the exposure of CL1-0 cells to 2-, 5-, and 10-Gy radiation reduced survival rates to 80%, 48.71%, and 17.32%, respectively. However, the exposure of CL1-5 cells to the same doses of radiation reduced their survival rates more significantly to 71%, 20.58%, and 2.44%, respectively (Figure 1). This result demonstrates that CL1-5 cells are more sensitive than CL1-0 cells to irradiation.
1H-NMR analysis for CL1-0 and CL1-5 at different time points after radiation exposure
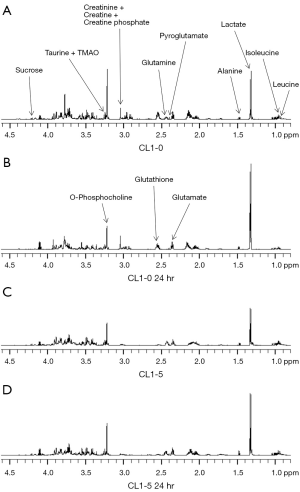
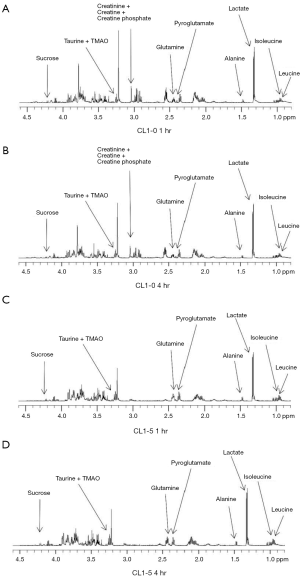
Our colony formation assay results showed that the surviving fraction of CL1-0 was dramatically larger than that of CL1-5 cells post 10-Gy exposure. Therefore, the CL1-0 and CL1-5 cells were treated with 10 Gy and harvested at 1, 4, and 24 hr to identify metabolites at different time points after 10-Gy exposure. The cell extracts were collected and evaluated using 1H-NMR. 1H-NMR spectra of representative extracts from CL1-0 and CL1-5 cells at 0, 1, 4, and 24 hr after irradiation treatment are shown in Figure 2 (0 and 24 hr) and Figure S1 (1 and 4 hr). Approximately 240 signals were detected by 1H-NMR. Of these signals, approximately 195 were concentrated in the range from 0 to 4.5 ppm. Therefore, 1H-NMR spectra in the range of 0 to 4.5 ppm were subjected to PCA analysis.
Multiparametric statistical analysis for CL1-0 and CL1-5 after radiation exposure
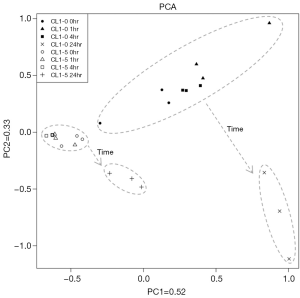
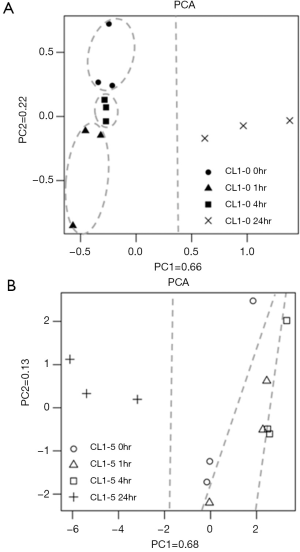
The PCA score plots for NMR spectra of CL1-0 and CL1-5 cells with or without radiation treatment are shown in Figure 3. The first two PCs capture a high proportion (85%) of the total variability, and a clear separation of the two cell lines can be seen. The clear separation of the two cell lines at the 0 hr time point indicates that the metabolite profiles of these two cell lines are inherently different. Moreover, the CL1-0 and CL1-5 cells follow similar trajectories from the upper left to the bottom right region of the plot from 0–4 to 24 hr.
Identification of metabolites in CL1-0 and CL1-5 with different radio-sensitivities
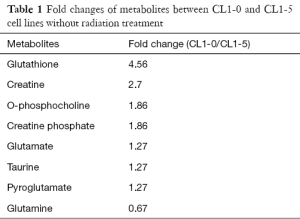
Since CL1-0 and CL1-5 cells presented inherently different metabolite profiles, the loading plots from the PCA plot were used to obtain the ppm value for further identification of specific metabolites from these two cell lines (Figure 4). The loading profile of PC1 explains 52% of the total variation in the spectra (Figure 3). The result shows that differences in relative concentrations of leucine, isoleucine, lactate, alanine, glutathione, glutamate, pyroglutamate, glutamine, creatinine, creatine, creatine phosphate, o-phosphocholine, taurine, trimethylamine n-oxide (TMAO), glucose, and sucrose are the main reasons for the appearance of the clusters. In addition to identifying the metabolites in the two cell lines, the fold changes of the differentially expressed metabolites were also measured. The fold change was calculated by dividing the median concentration in the CL1-0 cell line by that in the CL1-5 cell line. Table 1 shows the metabolites with greater than 1.25-fold changes between the two cell lines before irradiation. As shown in Table 1, the glutathione concentration in the CL1-0 cells was found to be 4.56-fold higher than in the CL1-5 cells. The minus sign indicates that the glutamine concentration in the CL1-0 cells is lower than in the CL1-5 cells.

Full table
PCA analysis of H-NMR data for CL1-0 and CL1-5 at different time points after radiation exposure
In order to identify the specific metabolite changes in response to radiation in CL1-0 or CL1-5 cells at different time points, the individual PCA score plots were further analyzed. In CL1-0 cells, the first two PCs capture a high proportion (88%) of the total variability, and each group of CL1-0 cells has its own specific scores along the PC2 direction that were gathered into a cluster (Figure 5A). The PCA score plots revealed that the clusters at 0, 1 and 4 hr time points were close to each other, indicating similarity at these times, while they were farther from the 24 hr point, indicating a large change at 24 hr following radiation. The PCA score plot for the CL1-5 cells is shown in Figure 5B. The first two PCs captured a high proportion (81%) of the total variability. The metabolite profiles of the CL1-5 cells changed with time after irradiation, and each time point was distributed along the PC1 axis.
Temporal dynamics of metabolite profiles
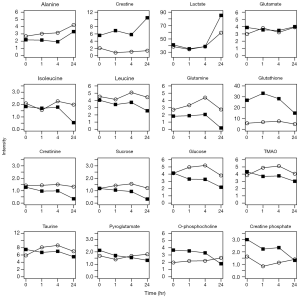
To better visualize the time-related changes for each metabolite, the changes in concentration of the sixteen metabolites at each time point in the CL1-0 and CL1-5 cells is shown in Figure 6. Several metabolite concentration profiles were changed similarly at the indicated time points after irradiation between the two cell lines including alanine, glutamine, isoleucine, lactate, leucine, sucrose, and glucose. Conversely, the metabolite concentration profiles of creatine, creatinine, creatine phosphate, glutamate, glutathione, o-phosphocholine, pyroglutamate, taurine, and TMAO were distinct between the two cell lines. The temporal dynamics of the metabolite concentrations uncover the different responses of the two cell lines upon irradiation.
Discussion
In this study, we aimed to identify distinct metabolic changes in human lung cancer cells with differential radiation sensitivities. We observed that the creatine phosphate, glutamate, glutathione, o-phosphocholine, pyroglutamate, taurine, and TMAO concentration profiles at 24 hr after irradiation are different in the two cell lines. Among these metabolites, the glutathione concentration in CL1-0 cells was inherently higher than in CL1-5 cells, and its concentration was elevated to a greater extent in CL1-0 cells compared to CL1-5 cells over time after 10-Gy irradiation. Some cancer cells with higher levels of glutathione are more drug-resistant to anticancer therapies (23). Glutathione has also been identified as a radio-protector as it removes free radicals, donating hydrogen to the damaged DNA radicals; reduces peroxides; and maintains protein thiols in the reduced state (24). Therefore, it is not surprising that radioresistant cells have higher glutathione levels compared to radiosensitive cells (25,26). After cells were irradiated, reactive oxygen species (ROS) were generated and accumulated within cells to induce cytotoxicity. Glutathione, as a ROS scavenger, detoxifies ROS by either spontaneous reduction or glutathione peroxidase-mediated reduction to reduce ROS induced cell death (27-31). Furthermore, targeting glutamine-dependent antioxidant capacity or glutathione metabolism increased cell death in radio-resistant cells (32,33). Therefore, the higher concentration of glutathione in CL1-0, as compared to CL1-5, might protect this cell line from ROS induced damage after radiation exposure. In addition to addressing the importance of glutathione in radiation treated CL1-0 and CL1-5 cells, the changes in other metabolites after radiation in these two lung cancer cells were examined to address the potential mechanism of radio-resistance in lung cancer.
As shown in Figure 6, the concentration changes in the indicated metabolites were similar in CL1-0 and CL1-5 cells over time after radiation exposure. Our PCA plot analysis also showed that there is a similar trajectory in PCA plots from the upper left to the bottom right region between 0–4 and 24 hr. Therefore, it is reasonable to assume that common metabolites might undergo alterations in these cells up to 24 hr after 10-Gy exposure; however, these metabolites require further investigation. In addition, glutathione concentration changed significantly with time in CL1-0 cells, while it remained unchanged in CL1-5 cells. In CL1-0 cells, the concentration of glutathione slightly increased soon after exposure to radiation and gradually decreased up to 24 hr after irradiation (Figure 6). This observation is consistent with previous studies, which have indicated increased endogenous glutathione levels following treatment with ionizing radiation (34,35). The change in glutathione concentration indicates a protective effect in cells. Oxidative stress results in the formation of glutathione disulfide (the oxidized form) at the expense of glutathione (reduced form). Glutamine and glutamate are precursors of glutathione, and their depletion in the cells is due to the synthesis of glutathione, which counteracts the effects of irradiation (36). Glutamate levels slightly decreased in the first 4 hr (Figure 6), possibly as a result of the consumption of glutamate to synthesize glutathione.
In this study, the concentration of taurine (the end product of cysteine catabolism) gradually declined after radiation treatment in CL1-0 cells (Figure 6). The decrease in taurine was also observed in other lung cancer cells after irradiation (27). The change in taurine concentration was different in CL1-5 cells; its concentration increased in the first 4 hr. This change could be explained by the osmoregulatory function of taurine (37). Because irradiation would destroy the cell membrane and the osmolytes, an increase in the taurine concentration may be necessary to balance the osmotic pressure. A previous study identified that taurine may have radioprotective effects (38). However, the slightly increased level of taurine in CL1-5 over time post radiation might not be sufficient for protection against radiation-induced damage. TMAO is involved in the detoxification process in the human body (39,40). The concentration profiles of TMAO were similar to those of taurine in both cell lines. This minor increase of TMAO concentration over time after radiation in CL1-5 also might be not enough to combat damage induced by ionizing radiation.
Lactate is the end product of glycolysis (41). Since 1953, investigators have observed that proliferating tumor cells can elevate the concentration of lactate at the expense of glucose (42,43). A high glycolytic rate is required to support tumor cell proliferation and duplication of the cell biomass and genome at each cell division. Lactate was recently identified as a major energy source in tumors (44). Moreover, lactate levels are significantly higher in lung cancer tissue than in normal lung tissue (45). In this study, the concentration of lactate in CL1-0 cells at 24 hr was significantly higher than in CL1-5 cells (Figure 6). In the meantime, the changes in the concentration profile of glucose also revealed that CL1-0 cells had a higher rate of glucose consumption, which may result in a higher survival rate of CL1-0 cells after irradiation.
The activity of creatine kinases has been shown to decrease progressively in sarcoma, and S180 cells in comparison with normal muscle (46), which suggests that cancer cell metabolism favors polyamine synthesis to meet the high energy demand of proliferating cancer cells (46). Compared with CL1-5 cells, the changes in concentrations of creatine and creatine phosphate in CL1-0 cells were more significant following radiation exposure. This phenomenon might contribute to the resistant nature of CL1-0 cells, which require more polyamine for better survival of cancer cells after irradiation.
Conclusions
Using NMR-based metabolomics analysis, we successfully identified specific metabolites contributing to the radio-sensitivity of human lung cancer cells. The primary metabolite that determines the radio-sensitivity of cells is glutathione, which could protect cells against oxidative stress as it has antioxidant properties. The higher endogenous levels in CL1-0 cells of metabolites such as glutathione, creatine phosphate, glutamate, o-phosphocholine, pyroglutamate, taurine, and TMAO were identified as potential key molecules responses to protecting radiation induced damage. Moreover, the changed concentration profiles of metabolites over time after radiation can be used to explain a possible mechanism of radio-resistance. The present study provides a starting point to investigate biomarkers for the radio-sensitivity of lung cancer. With the use of clinical samples with distinct responses to radiotherapy, specific biomarkers for radio-resistance could be verified, which could contribute to personalized medicine.
Acknowledgments
Funding: This study was sponsored by the College of Medicine (Aim for Top University Program), Ministry of Science and Technology, Taiwan (103-2314-B-002-034-MY3), and Electronics Engineering and Computer Science (Excellent Research Project), National Taiwan University.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.11.71). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Siegel R, Ward E, et al. Cancer Statistics, 2009. CA Cancer J Clin 2009;59:225-49. [Crossref] [PubMed]
- Toschi L, Cappuzzo F. Impact of biomarkers on non-small cell lung cancer treatment. Target Oncol 2010;5:5-17. [Crossref] [PubMed]
- Linden J. Optimizing radiation for cancer immunotherapy. Transl Cancer Res 2016;5:191-3. [Crossref]
- Rossi CJ Jr. Proton beam radiation therapy of prostate cancer—history, results, and future directions. Transl Cancer Res 2012;1:173-83.
- Bernard ME, Clump DA, LaLonde R, et al. Radiation therapy for locally advanced lung cancer. Transl Cancer Res 2015;4:356-71.
- Bayman N, Alam N, Faivre-Finn C. Radiotherapy for lung cancer in the elderly. Lung Cancer 2010;68:129-36. [Crossref] [PubMed]
- Repetto L, Balducci L. A case for geriatric oncology. Lancet Oncol 2002;3:289-97. [Crossref] [PubMed]
- Provencio M, Sanchez A, Garrido P, et al. New molecular targeted therapies integrated with radiation therapy in lung cancer. Clin Lung Cancer 2010;11:91-7. [Crossref] [PubMed]
- Nicholson JK, Connelly J, Lindon JC, et al. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discovery 2002;1:153-61. [Crossref] [PubMed]
- Yin C, Qie S, Sang N. Carbon source metabolism and its regulation in cancer cells. Crit Rev Eukaryot Gene Expr 2012;22:17-35. [Crossref] [PubMed]
- Nakajima T. Roles of Sulfur Metabolism and Rhodanese in Detoxification and Anti-Oxidative Stress Functions in the Liver: Responses to Radiation Exposure. Med Sci Monit 2015;21:1721-5. [Crossref] [PubMed]
- Souza-Filho JN, Guarnieri-Cruz MC, Murata Y, et al. Detoxification of the crotoxin complex by gamma radiation. Braz J Med Biol Res 1992;25:103-13. [PubMed]
- Zhirnov VV, Iakovenko IN. The osmotic resistance, and zeta potential responses of human erythrocytes to transmembrane modification of Ca2+ fluxes in the presence of the imposed low rate radiation field of 90Sr. Int J Radiat Biol 2015;91:117-26. [Crossref] [PubMed]
- Eo MY, Fan H, Cho YJ, et al. Cellulose membrane as a biomaterial: from hydrolysis to depolymerization with electron beam. Biomater Res 2016;20:16. [Crossref] [PubMed]
- Chu YW, Yang PC, Yang SC, et al. Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am J Respir Cell Mol Biol 1997;17:353-60. [Crossref] [PubMed]
- Chen JJ, Peck K, Hong TM, et al. Global analysis of gene expression in invasion by a lung cancer model. Cancer Res 2001;61:5223-30. [PubMed]
- Kang Y, Chang YY, Liu YC, et al. A microarray study of radiation-induced transcriptional responses and the role of Jagged 1 in two closely-related lung cancer cell lines. Transl Cancer Res 2015;4:314-23.
- Wold S, Esbensen K, Geladi P. Principal component analysis. Chemom Intell Lab Syst 1987;2:37-52. [Crossref]
- Madsen R, Lundstedt T, Trygg J. Chemometrics in metabolomics—A review in human disease diagnosis. Anal Chim Acta 2010;659:23-33. [Crossref] [PubMed]
- Lindon JC, Holmes E, Nicholson JK. Pattern recognition methods and applications in biomedical magnetic resonance. Progress in Nuclear Magnetic Resonance Spectroscopy 2001;39:1-40. [Crossref]
- Edlund U, Grahn H. Multivariate data analysis of NMR data. J Pharm Biomed Anal 1991;9:655-8. [Crossref] [PubMed]
- Liu YJ, Lin YF, Chen YF, et al. MicroRNA-449a enhances radiosensitivity in CL1-0 lung adenocarcinoma cells. PLoS One 2013;8:e62383 [Crossref] [PubMed]
- Tew KD. Glutathione-associated enzymes in anticancer drug resistance. Cancer Res 2016;76:7-9. [Crossref] [PubMed]
- Bump EA, Brown JM. Role of glutathione in the radiation response of mammalian cells in vitro and in vivo. Pharmacol Ther 1990;47:117-36. [Crossref] [PubMed]
- Lu TP, Lai LC, Lin BI, et al. Distinct signaling pathways after higher or lower doses of radiation in three closely related human lymphoblast cell lines. Int J Radiat Oncol Biol Phys 2010;76:212-9. [Crossref] [PubMed]
- Yang P, Ebbert JO, Sun Z, et al. Role of the glutathione metabolic pathway in lung cancer treatment and prognosis: a review. J Clin Oncol 2006;24:1761-9. [Crossref] [PubMed]
- Rainaldi G, Romano R, Indovina P, et al. Metabolomics using H-1-NMR of apoptosis and necrosis in HL60 leukemia cells: Differences between the two types of cell death and independence from the stimulus of apoptosis used. Radiation Res 2008;169:170-80. [Crossref] [PubMed]
- Anderson ME, Luo JL. Glutathione therapy: From prodrugs to genes. Semin Liver Dis 1998;18:415-24. [Crossref] [PubMed]
- Traverso N, Ricciarelli R, Nitti M, et al. Role of glutathione in cancer progression and chemoresistance. Oxid Med Cell Longev 2013;2013:972913.
- Chiang YT, Yen YW, Lo CL. Reactive oxygen species and glutathione dual redox-responsive micelles for selective cytotoxicity of cancer. Biomaterials 2015;61:150-61. [Crossref] [PubMed]
- Oien DB, Chien J, Cheng N. Regulation of chemo-sensitivity in ovarian cancer via a stroma dependent glutathione pathway. Transl Cancer Res 2016;5:S514-9. [Crossref]
- Matschke J, Riffkin H, Klein D, et al. Targeted inhibition of glutamine-dependent glutathione metabolism overcomes death resistance induced by chronic cycling hypoxia. Antioxid Redox Signal 2016;25:89-107. [Crossref] [PubMed]
- Mena S, Rodriguez ML, Ortega A, et al. Glutathione and Bcl-2 targeting facilitates elimination by chemoradiotherapy of human A375 melanoma xenografts overexpressing bcl-xl, bcl-2, and mcl-1. J Transl Med 2012;10:8. [Crossref] [PubMed]
- Kojima S, Matsumori S, Ono H, et al. Elevation of glutathione in RAW 264.7 cells by low-dose gamma-ray irradiation and its responsibility for the appearance of radioresistance. Anticancer Res 1999;19:5271-5. [PubMed]
- Pujari G, Berni A, Palitti F, et al. Influence of glutathione levels on radiation-induced chromosomal DNA damage and repair in human peripheral lymphocytes. Mutat Res 2009;675:23-8. [Crossref] [PubMed]
- Chatterjee A. Reduced glutathione: a radioprotector or a modulator of DNA-repair activity? Nutrients 2013;5:525-42. [Crossref] [PubMed]
- Rockel N, Esser C, Grether-Beck S, et al. The osmolyte taurine protects against ultraviolet B radiation-induced immunosuppression. J Immunol 2007;179:3604-12. [Crossref] [PubMed]
- Christophersen OA. Radiation protection following nuclear power accidents: a survey of putative mechanisms involved in the radioprotective actions of taurine during and after radiation exposure. Microb Ecol Health Dis 2012;23. [PubMed]
- Dos Santos JP, Iobbi-Nivol C, Couillault C, et al. Molecular analysis of the trimethylamine N-oxide (TMAO) reductase respiratory system from a Shewanella species. J Mol Biol 1998;284:421-33. [Crossref] [PubMed]
- Athawale MV, Dordick JS, Garde S. Osmolyte trimethylamine-N-oxide does not affect the strength of hydrophobic interactions: Origin of osmolyte compatibility. Biophys J 2005;89:858-66. [Crossref] [PubMed]
- Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nat Rev Cancer 2004;4:551-61. [Crossref] [PubMed]
- Warburg O. On respiratory impairment in cancer cells. Science 1956;124:269-70. [PubMed]
- Warburg O. Origin of cancer cells. Science 1956;123:309-14. [Crossref] [PubMed]
- Feron O. Pyruvate into lactate and back: From the Warburg effect to symbiotic energy fuel exchange in cancer cells. Radiother Oncol 2009;92:329-33. [Crossref] [PubMed]
- Yokota H, Guo JF, Matoba M, et al. Lactate, choline, and creatine levels measured by vitro H-1-MRS as prognostic parameters in patients with non-small-cell lung cancer. J Magn Reson Imaging 2007;25:992-9. [Crossref] [PubMed]
- Bera S, Wallimann T, Ray S, et al. Enzymes of creatine biosynthesis, arginine and methionine metabolism in normal and malignant cells. FEBS J 2008;275:5899-909. [Crossref] [PubMed]


