
JMJD3 histone demethylase—a potential therapeutic target for metastatic hepatocellular carcinoma?
Hepatocellular carcinoma (HCC) remains as a top cause of cancer death worldwide. One of challenges in the history of finding ways to eradicate HCC is its complicated pathobiology and its high potential of metastasis, which is not fully understood in terms of mechanisms. Particularly, the mechanism by which metastasis is associated with the hallmarks of cancer, such as aberrant epigenetics modification and genome instability, is still far from clear. The link among all these malignant behaviors and the root of cancer recurrence, the cancer stemness, is even more unclear.
A recent paper published in Cancer Research provides some insight into the association of histone methylation, metastasis and cancer stemness (1). Tang et al. described the upregulation of JMJD3 in metastatic HCC while many other Jumonji C (JmjC) domain-containing factors were not altered. JMJD3 overexpression was negatively associated with patient survival. Using cellular model of stable overexpression and knockdown, they systematically showed that JMJD3 expression favors cell proliferation and tumor formation. The upregulation of JMJD3 also facilitates cell migration and invasion by promoting epithelial-mesenchymal transition (EMT). More importantly, JMJD3 expression was shown to enhance the stem cell-like behaviors in HCC cell-lines while overexpression of JMJD3 was validated in cancer stem cell markers, EpCam- and CD133-positive HCC cells. Through screening, they found that Slug gene was positively regulated by JMJD3 through histone demethylation at the promoter region.
JMJD3 belongs to a large family of histone demethylases containing JmjC domains. Different members of the family have different specificity on histone demethylation. For example, JMJD6 is a histone arginine demethylase targeting H3 or H4 (2). In contrast, JMJD3 demethylates H3K27 as an action to revert polycomb group (PcG)-mediated lysine methylation (3). Lysine methylation can occur in mono, di- or trimethylated forms, in which they influence the chromatin structure and therefore gene expression in either direction (3). Thanks to the advancement in deep sequencing technology which helps to reveal the general association of H3K27 trimethylation with repressed promoters, whereas H3K3 di- and trimethylation are highly abundant in active promoters (4). In other words, the abundance in various types of histone methylation determines the general overall gene expression patterns. In human, H3K27 trimethylation is executed by a complex called Polycomb Repressive Complex 2 (PRC2) which is composed of the enzyme EZH2, FED and SUZ12 (5). On the other hand, UTX (ubiquitously transcribed tetratricopeptide repeat, X chromosome) and JMJD3 specifically remove methylation marks from EZH2-methylated H3K27m3 (6) (Interested readers are referred to the review written by Varier in 2001.). Thus, the expression of JMJD3 represents a shift of the equilibrium towards histone demethylation, and thereby the general re-expression of otherwise silenced genes (Figure 1).
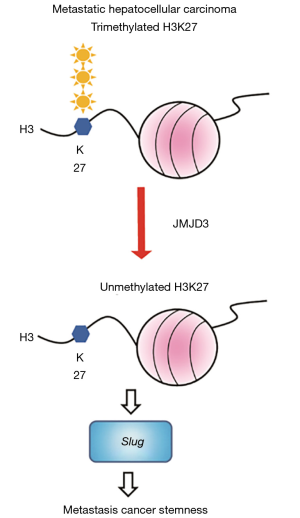
The oncogenic role of JMJD3 has been documented in the past five years. This work represents the first evidence on the contribution of JMJD3 in metastatic phenotypes in HCC. For years, efforts have been made to explore the genes or chromosomes associated with HCC metastasis. As summarized by Tang et al., several genes were now proven to be linked with HCC metastasis including p16, p21, p53, TGFα, EGFR, c-erbB-2, nm23-H1 and TIMP-2 (7). These genes may be mutated or deregulated. Recently, our group also connected phosphorylation to metastasis as PAK4 can phosphorylate p53 to promote HCC metastasis (8). Aberrant histone methylation has been implicated in HCC, where the level of H3K4me2 is relatively low (9). The new findings described by Tang et al. further enhances our understanding on the epigenetic regulation of HCC metastasis, bringing in a new speculation that the level of H3K27m3 may be generally low in HCC. However, this is subject to further study that specifically determines the pattern of H3K27me3 and to profile the genes re-expressed by JMJD3. It was found that H3K27 trimethylation pre-marks the packaged genes for de novo DNA methylation (10). This implies a more complicated epigenetic regulation that may be brought by aberrant JMJD3 expression. Hence, we believe there is a great potential for further investigation of JMJD3-associated epigenetic regulation in HCC.
The work by Tang et al. also showed that epithelial-to-mesenchymal transition gene Slug is highly expressed through H3K27 demethylation by JMJD3. In recent years, considerable amount of evidence showed that cells undergo EMT displayed cancer stem cell features and vice versa (11). In line with this, the authors also showed that JMJD3 is enriched in CD133+ HCC stem cells, indicating that H3K27 histone demethylation may underlie the aberrant expression of many genes in maintaining the cancer stemness. However, it is likely yet unclear whether JMJD3 upregulation is essential in maintaining cancer stemness rather than a “product” of stemness, as stable expression JMJD3 in HCC cells exhibits stemness properties. Cancer stem cells has the property of self-renewal and the capacity to differentiate into multiple lineages of cells (12). Several pathways such as Notch, Wnt, Hedgehog are known to be actively involved in cancer stem cells (11). Thus, a regulation of these pathways by JMJD3 remains an interesting issue to be addressed.
Furthermore, it will be insightful to study the detailed consequences of histone demethylation in cancer stem cells. Because of the reversible nature of epigenetic regulation, it is plausible that histone methylation may act as an alternative approach to suppress cancer stemness and metastasis (13). As a conclusion, Tang et al. suggested the reversion of JMJD3 expression via suppression of its enzymatic activity as a potential HCC therapeutic option. The resolve of the ligand-bound structure of human JMJD3 should make this approach more feasible. In 2012, Kruidenier et al. described a structure-based identification of small molecules that bind to the catalytic pocket of JMJD3 (14). Among them, GSK-J1 was found effective in inhibiting the activity of JMJD3 using cell culture assays. Since GSK-J1 biomolecule is readily available from the commercial market, it will be of great interest to investigate if GSK-J1 suppresses the activity of JMJD3 and reduces malignant features of HCC cells. Equally important is to study how inhibition of JMJD3 will alter other genes expressions in a genome-wide manner.
In conclusion, the work presented by Tang et al. has widened the horizon on understanding HCC metastasis. We believe such finding will lead us to an exciting path of exploring epigenetic regulation in HCC metastasis and in stemness of cancer cells.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Bo Zhai (Department of Hepatobiliary Surgery, The Fourth Hospital of Harbin Medical University, Harbin, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.12.61). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tang B, Qi G, Tang F, et al. Aberrant JMJD3 expression upregulates slug to promote migration, invasion, and stem cell-like behaviors in hepatocellular carcinoma. Cancer Res 2016; [Epub ahead of print]. [Crossref] [PubMed]
- Chang B, Chen Y, Zhao Y, et al. JMJD6 is a histone arginine demethylase. Science 2007;318:444-7. [Crossref] [PubMed]
- Varier RA, Timmers HT. Histone lysine methylation and demethylation pathways in cancer. Biochim Biophys Acta 2011;1815:75-89.
- Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell 2007;129:823-37. [Crossref] [PubMed]
- Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev 2004;14:155-64. [Crossref] [PubMed]
- Hong S, Cho YW, Yu LR, et al. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A 2007;104:18439-44. [Crossref] [PubMed]
- Tang ZY, Ye SL, Liu YK, et al. A decade's studies on metastasis of hepatocellular carcinoma. J Cancer Res Clin Oncol 2004;130:187-96. [Crossref] [PubMed]
- Xu HT, Lai WL, Liu HF, et al. PAK4 phosphorylates p53 at serine 215 to promote liver cancer metastasis. Cancer Res 2016;76:5732-42. [Crossref] [PubMed]
- Magerl C, Ellinger J, Braunschweig T, et al. H3K4 dimethylation in hepatocellular carcinoma is rare compared with other hepatobiliary and gastrointestinal carcinomas and correlates with expression of the methylase Ash2 and the demethylase LSD1. Hum Pathol 2010;41:181-9. [Crossref] [PubMed]
- Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 2007;39:232-6. [Crossref] [PubMed]
- Książkiewicz M, Markiewicz A, Zaczek AJ. Epithelial-mesenchymal transition: a hallmark in metastasis formation linking circulating tumor cells and cancer stem cells. Pathobiology 2012;79:195-208. [Crossref] [PubMed]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer 2008;8:755-68. [Crossref] [PubMed]
- Monteiro J, Fodde R. Cancer stemness and metastasis: therapeutic consequences and perspectives. Eur J Cancer 2010;46:1198-203. [Crossref] [PubMed]
- Kruidenier L, Chung CW, Cheng Z, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 2012;488:404-8. [Crossref] [PubMed]


