
Response letter: “epigenetic modulation of a miR-296-5p:HMGA1 axis regulates Sox2 expression and glioblastoma stem cells”
In this issue of Translational Cancer Research Ferrucci and Zollo provide a thoughtful commentary discussing our recent Oncogene publication titled “Epigenetic Modulation of a miR-296-5p:HMGA1 Axis Regulates Sox2 expression and Glioblastoma Stem Cells”. The experts examined our results with a critical eye and comment on the strengths and weaknesses of our study while at the same time providing very thoughtful and considerate suggestions for future studies and new directions.
We share with Ferrucci and Zollo the view that most solid tumors, including GBM, contain sub-sets of cells that display stem-like qualities. These so called “cancer stem cells” (or CSCs) play a key role in maintaining tumor growth, contribute to resistance, and drive tumor recurrence (1). In recent years our laboratory demonstrated that oncogenic receptor tyrosine kinase signaling drives the glioma stem cell phenotype by inducing reprogramming transcription factors and that inhibiting this axis in vivo depletes tumors of their stem-like tumor-propagating cells (2,3). We also found that co-expressing the reprogramming transcription factors Oct4 and Sox2 in tumor cells that lack stemness and tumor-propagating potential induces de-differentiation and, more significantly, tumor propagating capacity (4). These reprogramming events were found to involve the direct activation of DNA methyl-transferase gene transcription, increased DNA methylation, and methylation-dependent repression of miRNA networks containing miRNAs that inhibit tumor cell stemness and tumor propagating potential (4,5).
We now realize that the CSC pools are very dynamic and they can gain or lose the stem-like tumor-propagating phenotype (i.e., “stemness”) depending on contextual molecular cues resulting in heterogeneous populations of tumor cells that can either expand in a limited fashion (i.e., transit amplifying cells) or actually propagate tumors (i.e., CSCs). We are now starting to understand that this phenotypic heterogeneity is supported by reprogramming molecular circuits and networks that function within the context of an aberrant cancer genome. We share the overall view that the mechanisms driving these phenotypic transitions represent a vulnerability amenable to therapeutic targeting.
In our earlier study we describe how these reprogramming events in GBM are driven by Oct4 and Sox2 and involve activation of DNA methyl-transferase (DNMT) gene transcription and methylation-dependent repression of stemness inhibiting miRNAs focusing more specifically on miR-148a (4). We show in the current complementary study that this process also involves the repression of miRNA 296-5p that in turn targets the chromatin architectural transcriptional regulator HMGA1 feeding back upon Sox2 expression (5). Ferrucci and Zollo correctly point out that Sox2 is highly enriched in CD133+ GBM stem-like cells (6) and several lines of evidence including our findings indicate that Sox2 is required to maintain the stem cell phenotype in different contexts (7). Ferrucci and Zollo propose a new therapeutic paradigm involving Sox2 inhibition in combination with drugs that target reprogramming epigenetic mechanism. We agree that this is a logical direction for therapeutic development with the caveat that transcription factors are not especially “druggable” and that pharmacologic modulation of epigenetic enzymes may lack sufficient biological specificity. In fact, several drugs that target epigenetic mechanism, including 5-azacytidine to target DNMTs, have been developed and tested in the clinic with limited success (8). Reconstituting molecular networks associated with more differentiated transit-amplifying cancer cells and absent in tumor-propagating CSCs represents a highly rational albeit challenging alternative approach. We have described two miRNAs (miR-148a and miR-296-5p) whose reconstitution in conjunction with other molecular modulations could be used to achieve such a goal (4,5). In addition to DNMT-dependent repression of miRNAs that inhibit GBM CSCs, Oct4 and Sox2 concurrently induce miRNAs that act as “onco-miRs” {e.g., miR-10b (4,9)}. Suppressing these CSC-inducing oncogenic miRNAs in combination with restoring CSC-inhibiting tumor-suppressor miRNAs will likely be necessary. Nonviral nucleic acid delivery vehicles formulated specifically for miRNA delivery offer a promising technology for delivering combinations of miRNAs and antago-miRs to test these concepts in pre-clinical models and for clinical translation (10).
Ferrucci and Zollo present an interesting bioinformatics analysis to predict protein networks regulated by miR-296-5p targets highlighting prominent effects on differentiating signals. However, we advise caution in applying these results to GBM CSCs since the predictions are based on protein activities identified in different tumor types and cellular contexts. We also wish to point out that even though restoring miR-296-5p decreased stem cell drivers, stem cell markers and neurosphere self-renewal in our experimental systems, we never observed a change in the expression of neural lineage markers such as GFAP, Tuj-1, or O4 as observed following the reconstitution of miR-148a (4). This suggests that miR-296-5p inhibits self-renewal capacity but is not sufficient to drive GBM CSC differentiation. One possibility is that miR-296-5p functions to lower the threshold for differentiation, an activity that could cooperate with other molecular or pharmacologic differentiation signals such as miR-148a or retinoic acid. We echo Ferrucci and Zollo’s interest in these promising directions and believe that differentiation therapies to force tumor cells into phenotypes that are less efficient tumor propagators and/or more susceptible to treatment is an attractive therapeutic strategy.
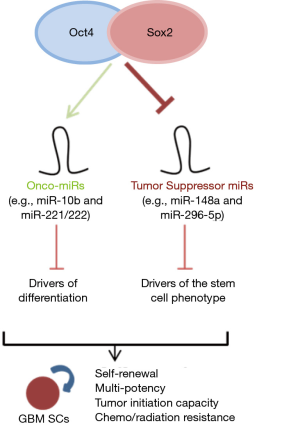
Our results so far have established a working model in which Oct4/Sox2 drive glioma stem cell formation by differentially regulating a network of miRNAs. Our newfound mechanisms show that the combined action of Oct4/Sox2 induce glioma cell stemness and tumor-propagating potential by simultaneously activating onco-miRs {e.g., miR-10b (4,10) } and repressing tumor suppressing miRs (e.g., miR-148a and miR-296-5p). The coordinate action of these two parallel pathways leads to inhibition of drivers of differentiation and activation of drivers of the stem cell phenotype ultimately resulting in signals that drive neoplastic cell stemness, self-renewal, and tumor propagating potential (Figure 1). Identifying the components and circuitries that generate and maintain CSCs and finding new ways of targeting them will allow us to design rational molecular therapies aimed at inhibiting the phenotypic states that drive tumor growth and recurrence.
Acknowledgments
Funding: This work was supported by grants from the American Brain Tumor Association (YL and HLB), and the United States NIH grants R01NS073611 (JL) and R01NS070024 (AQ-H).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Lichao Sun (State Key Laboratory of Molecular Oncology, National Cancer Center (NCC) /Cancer Hospital, Chinese Academy of Medical Sciences (CAMS), Peking Union Medical College, Beijing, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2016.12.29). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lopez-Bertoni H, Li Y, Laterra J. Cancer Stem Cells: Dynamic Entities in an Ever-Evolving Paradigm. Biol Med (Aligarh) 2015;7:001 [PubMed]
- Rath P, Lal B, Ajala O, et al. In Vivo c-Met Pathway Inhibition Depletes Human Glioma Xenografts of Tumor-Propagating Stem-Like Cells. Transl Oncol 2013;6:104-11. [Crossref] [PubMed]
- Li Y, Li A, Glas M, et al. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci U S A 2011;108:9951-6. [Crossref] [PubMed]
- Lopez-Bertoni H, Lal B, Li A, et al. DNMT-dependent suppression of microRNA regulates the induction of GBM tumor-propagating phenotype by Oct4 and Sox2. Oncogene 2015;34:3994-4004. [Crossref] [PubMed]
- Lopez-Bertoni H, Lal B, Michelson N, et al. Epigenetic modulation of a miR-296-5p:HMGA1 axis regulates Sox2 expression and glioblastoma stem cells. Oncogene 2016;35:4903-13. [Crossref] [PubMed]
- Song WS, Yang YP, Huang CS, et al. Sox2, a stemness gene, regulates tumor-initiating and drug-resistant properties in CD133-positive glioblastoma stem cells. J Chin Med Assoc 2016;79:538-45. [Crossref] [PubMed]
- Weina K, Utikal J. SOX2 and cancer: current research and its implications in the clinic. Clin Transl Med 2014;3:19. [Crossref] [PubMed]
- Di Costanzo A, Del Gaudio N, Migliaccio A, et al. Epigenetic drugs against cancer: an evolving landscape. Arch Toxicol 2014;88:1651-68. [Crossref] [PubMed]
- Ji Y, Wei Y, Wang J, et al. Correlation of microRNA-10b upregulation and poor prognosis in human gliomas. Tumour Biol 2015;36:6249-54. [Crossref] [PubMed]
- Tzeng SY, Green JJ. Therapeutic nanomedicine for brain cancer. Ther Deliv 2013;4:687-704. [Crossref] [PubMed]


