
DNA double strand break repair via non-homologous end-joining
Introduction
The human genome is constantly under assault from agents generated from both inside and outside of the cell. These assaults of the genome result in the generation of tens of thousands of DNA lesions per day in each human, with the most deleterious of these lesions being the DNA double strand break (DSB) (1). Introduction of a DSB can be mediated by a variety of means including those by endogenous sources like reactive oxygen species generated by cellular metabolism and replication associated errors and exogenous sources including ionizing radiation and chemotherapeutic agents (2). DSBs are also intentionally generated during V(D)J recombination and class switch recombination for the development of T- and B-cells [reviewed (3)]. Unrepaired or misrepaired DSBs can result in senescence, inducted apoptosis, or chromosomal aberrations including translocations and deletions which can result in a loss of heterozygozity. These chromosomal aberrations are associated with genomic instability and can ultimately result in carcinogenesis; therefore, it is of the utmost importance that cells have a mechanism(s) to quickly repair DSBs. A number of highly efficient DSB repair pathways have evolved in eukaryotic cells with non-homologous end-joining (NHEJ) likely playing the largest role in DSB repair in humans (4). The NHEJ pathway is also essential for V(D)J recombination during T- and B-cell lymphocyte development (3).
NHEJ mediates the direct re-ligation of the broken DNA molecule (5). It has the potential to re-ligate any type of DNA ends and unlike the other classically studied DSB repair mechanism, homologous recombination (HR), NHEJ does not require a homologous template for repair of the DNA lesion. Since NHEJ does not require a homologous template, theoretically it is not restricted to a certain phase of the cell cycle, whereas HR is believed to be only active during S and G2 phases of the cell cycle when a homologous template via the sister chromatid is available. The general mechanism of NHEJ can be broken down into individual and sequential steps which are: (I) DNA end recognition and assembly and stabilization of the NHEJ complex at the DNA double strand break; (II) Bridging of the DNA ends and promotion of end stability; (III) DNA end processing; and (IV) Ligation of the broken ends and dissolution of the NHEJ complex. In this review we will discuss each step of the pathway and the interesting new insights into the mechanism of the NHEJ pathway and the proteins which mediate this repair process. Furthermore, the role of the kinase activity of the DNA dependent protein kinase catalytic subunit (DNA-PKcs) and its possible role(s) in NHEJ will be outlined. Finally, the general role of NHEJ in promoting genomic stability and its implications in cancer incidence will be discussed.
DNA end recognition and assembly and stabilization of the NHEJ complex at the DNA double strand break
The initial step in NHEJ is the recognition and binding of the Ku heterodimer to the DSB (6,7) (Figure 1A,B). The Ku heterodimer is composed of the Ku70 and Ku80 (also called Ku86) subunits. Although there is little sequence identity between Ku70 and Ku80, the two Ku subunits show similar domain organization with each subunit being composed of three domains: an amino-terminal von Willebrand domain (vWA), a central Ku core domain, and a divergent carboxyl-terminal region (8). The vWA and Ku core domains are involved in the heterodimerization of the complex. The carboxyl-terminal region of Ku70 contains a SAF-A/B, Acinus and PIAS (SAP) domain which is likely a DNA binding domain (9,10). The Ku80 C-terminus forms a flexible arm which resembles a common protein scaffold that is involved in protein-protein interactions (11). Ku70/80 has been shown to localize to laser-generated DSBs within seconds of their creation (6). The ability of Ku to quickly localize to DSBs is likely due to its extraordinary affinity (binding constant of 2×109 M-1) for DNA ends, its ability to interact with DSBs in a sequence independent manner, and its abundant concentration (~500,000 Ku molecules/cell) (8,12-14). Crystallographic studies of Ku70/80 show that the heterodimer produces a ring-shaped structure which can accommodate a double strand DNA helix and allows the Ku heterodimer to slide onto the DNA end (13). Furthermore, the crystal structure shows that Ku binds to the sugar backbone of DNA and not to the bases which explains the ability of Ku to bind to DNA in a sequence independent manner.
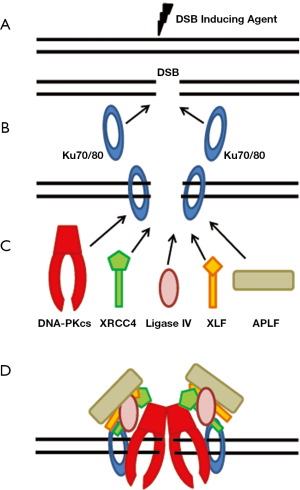
Once the Ku heterodimer is bound to the DSB ends, it then serves as a scaffold to recruit the other NHEJ factors to the damage site (Figure 1B,C). The Ku heterodimer has been shown to recruit either directly or indirectly the main NHEJ factors, including DNA-PKcs (7), X-ray cross complementing protein 4 (XRCC4) (6,15,16), DNA Ligase IV (15), XRCC4-like factor (XLF) (17), and Aprataxin-and-PNK-like factor (APLF) (18-20) to DSBs (Figure 1C). Terminal deoxynucleotidyl transferase (TdT) associates directly with Ku, but TdT is only found in lymphocytes and thus likely only plays a role in NHEJ in V(D)J recombination (21). Much work has been performed to delineate the key domains/residues responsible for mediating the interaction between Ku and the other NHEJ factors. The most studied interaction is the one between Ku70/80 and DNA-PKcs. DNA-PKcs is a member of phosphatidylinositol-3 (PI-3) kinase-like kinase family (PIKK), which includes two other protein kinases that play important roles in the cellular response to DNA damage, ataxia-telangiectasia mutated (ATM) and (ATM and Rad3- related (ATR) (22,23). Structurally the N-terminal region of each member of the PIKK family is composed of HEAT (Huntington-elongation-A-subunit-TOR) repeats which likely serve as a protein-protein interaction interface (24). The N-terminal region of DNA-PKcs contains a leucine zipper which contributes to its innate DNA affinity (25). The C-terminal region of the family contains a significant amount of sequence similarity with the C-terminal region of each protein containing a FAT (FRAP, ATM, TRRAP), PI3 kinase, and a FATC (FAT C-terminal) domain (26). The FAT and FATC domains surround the kinase domain and likely play an important role in regulating kinase activity of the PIKK family members (27). Structural studies of DNA-PKcs show that the C-terminal domains form a crown structure with the N-terminal portion of the protein producing a pincer-shaped structure which forms a central channel that likely mediates the ability of DNA-PKcs to bind to dsDNA (28,29). Ku70/80 directly recruits DNA-PKcs to the DNA ends to form the active DNA-PK complex and the interaction between the two requires the presence of DNA (30). Binding of DNA-PKcs to the DNA-Ku complex results in translocation of the Ku heterodimer inward on the dsDNA strand and ultimately results in activation of the DNA-PKcs kinase activity. A number of studies implicated that the extreme C-terminal 12 residues of Ku80 mediated the recruitment of DNA-PKcs to DSB ends (31-33). However, the Ku80 carboxyl terminus is not required for the ability of DNA-PKcs to localize to laser-generated DSBs in vivo suggesting that there are likely multiple residues and/or motifs mediating the ability of DNA-PKcs to interact with the Ku-DNA complex (34). In vitro binding studies previously showed that Ku70/80 interacts with amino acids 3,002-3,850 in the C-terminal region of DNA-PKcs (35). Recent experimental evidence by our group shows that the N-terminal region of DNA-PKcs is required for the ability of DNA-PKcs to interact with the Ku-DNA complex (36). It is likely the central cavity formed by the N-terminal region of DNA-PKcs results in DNA threading through the channel and ultimately stabilization of the DNA-PKcs-Ku-DNA complex (28,29). Predictions from low resolution structure shows that Ku70/80 makes multiple contacts with DNA-PKcs including contacts with the N- and C-terminal regions of the protein; therefore, it is likely that the N- and C-terminal regions of the DNA-PKcs make contacts with Ku70/80, but that the N-terminal region is absolutely necessary for the ability of the protein to interact with and/or be stabilized by the Ku-DNA complex (37,38).
Ku physically interacts with the XRCC4-DNA Ligase IV complex and recruits it to DNA ends in vitro and in vivo (6,16). XRCC4 directly interacts with the Ku70 subunit of the Ku heterodimer (6) whereas DNA Ligase IV directly interacts with the Ku heterodimer, and this interaction is mediated by the tandem BRCA1 C-terminal (BRCT) domains found in C-terminus of DNA LigaseIV, in particular the first BRCT domain (amino acids 644-748) (15,39). XLF interacts with the Ku heterodimer in a DNA dependent manner and this interaction is mediated by the heterodimeric domain of Ku and the C-terminal region of XLF from amino acids 270-299 (17,40). Recently, it was found that a conserved peptide between residues 182-191 in the MID domain of APLF interacts directly with the vWA domain of Ku80 (18).
XRCC4 may be a second NHEJ scaffold responsible for the recruitment of a number of NHEJ factors to the DSB ends; in particular it may play a role in securing the ability of the processing enzymes to interact with the DSB region (Figure 2). XRCC4 has no known enzymatic activity and is composed of a globular head domain, an elongated alpha-helical stalk, and C-terminal tail (41). XRCC4 homodimerizes and two dimers can make tetramers. The best studied processing enzyme that XRCC4 interacts with is DNA ligase IV. DNA Ligase IV has a carboxyl-terminal tandem BRCT domain and the linker region between the two BRCT domains and second BRCT domain mediates the interaction between DNA Ligase IV and the central alpha-helical stalk of XRCC4 (42,43). DNA polymerase µ is stably recruited to DNA in the presence of both Ku and XRCC4-Ligase IV (44). Similarly, the RecQ helicase family member Werner (WRN) interacts with both Ku and XRCC4-Ligase IV suggesting that XRCC4 in conjunction with Ku can mediate the recruitment of processing enzymes to DSBs (45-47). The polynucleotide kinase-phosphatase (PNKP) interacts with XRCC4 via its forkhead-associated (FHA) domain (48). This interaction is dependent on casein kinase 2 (CK2) phosphorylation of XRCC4. PNKP is phosphorylated itself by ATM on serine 114 and on serine 126 by ATM and DNA-PKcs (49). Although phosphorylation at these sites does not positively or negatively affect the ability of PNKP to interact with XRCC4, PNKP recruitment to laser-generated DSBs is impaired in cells deficient for DNA-PKcs and ATM suggesting that phosphorylation of PNKP or a factor by DNA-PKcs and ATM play a role in the ability of PNKP to localize to DSBs. However, it was also shown that PNKP interacts with unphosphorylated XRCC4 through a lower affinity interactions site (50). APLF has been reported to have endo/exonuclease activity and it interacts with threonine 233 of XRCC4 in an FHA and phospho-dependent manner (19-20,51). Similar to PNKP, aprataxin is a protein which contains a FHA domain and interacts with XRCC4 in a CK2-phosphorylated manner via its FHA domain (52). Taken together, XRCC4’s role in NHEJ appears to be much more complex than just basic recruitment of the terminal ligase for the reaction to the DSB.
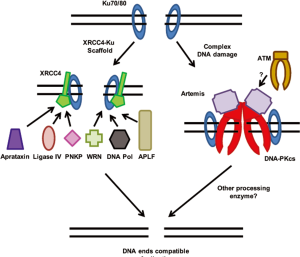
The prevailing model for NHEJ has been a sequential stepwise recruitment of each component or complex to the DSB. Following recruitment of Ku to the DSB, DNA-PKcs was believed to be first recruited to the DSB where its kinase activity is activated. Next, Ku along with DNA-PKcs recruited the processing enzymes to the DSB and lastly the XRCC4/Ligase IV complex was finally recruited to mediate ligation of the DSB. However, this sequential stepwise recruitment model may actually not be correct (53). There is no question that Ku is the first NHEJ factor to bind to the DSB end and it is required for the recruitment of all subsequent proteins to the DSB in vivo. However, DNA-PKcs is not necessarily the next recruited to the DSB and is not required for the recruitment of the other NHEJ factors to the DSBs as shown by the fact that localization of XRCC4, DNA Ligase IV, and XLF to DSB is not dependent on DNA-PKcs (6,53). The order of recruitment of the factors outside of the Ku heterodimer may actually be flexible and may depend on the complexity of the DNA damage (54). Simple DSBs may be repaired rapidly only involving the Ku heterodimer, XRCC4, LigaseIV, and XLF, whereas complex breaks requires DNA-PKcs and possibly the activity of ATM.
Finally, a number of factors have been believed to stabilize the NHEJ complex at DSBs including Ku, DNA-PKcs, the kinase activity of DNA-PKcs, and XRCC4 (55). Recently, it was shown that APLF may play a role in assembly of the NHEJ complex and that its main role may be to promote the retention of XRCC4, DNA LigaseIV, and XLF at DSBs (18,56). It seems likely that a multitude of protein-DNA and protein-protein interactions between the NHEJ factors stabilize the entire core complex at the DSB (Figure 1D). Taken together, the recent data suggests that there may be subcomplexes or subpathways of NHEJ that may mediate end-joining depending on the complexity and/or nature of the DSB. Ku has been implicated to function in a “tool belt manner” where it can recruit, depending on the manner of the DSB, whichever enzymatic activity that is required for repair of the DSB (57). It is likely that XRCC4 may also play a role in the ability of the NHEJ complex to choose the correct enzymes to aide in the repair of a specific break depending on the nature of the break and the ends. It will be of interest to tweeze out what factors are required for specific damages and which proteins play a role in stabilizing the repair proteins and required for the processing of the damage.
Bridging of the DNA ends and promotion of end stability
Upon the creation of a DSB, Ku70/80 binds to the DNA ends and helps maintain their stability by protecting them from non-specific processing (Figure 3A). This is of importance, as non-specific processing of ends could lead to chromosomal aberrations and thus genomic instability. Atomic force microscopy showed that Ku itself can hold the two termini of a linearized plasmid in a synaptic complex further suggesting that Ku can bind to and protect DNA ends (58). Utilizing a live cell imaging approach, it was shown that Ku80, but not the Mre11-Rad50-Nbs1 complex or the structural maintenance of chromosomes protein 1, is required for the positional stability of DSB ends in asynchronized mouse cells (59). Ku’s role of protecting the DSB ends may happen in all cell cycle phases as our group has shown that the accumulation and dissociation kinetics of Ku80 at micro-laser induced DSBs are independent of cell cycle (60). Furthermore, it has been reported that Ku deficient cells have severe chromosome instability when these cells were irradiated in S phase (61). Taken together, these data suggest that Ku70/80 plays a role at DSB ends even when HR is the preferred repair pathway. Once Ku recruits DNA-PKcs to the DSB ends, it has been shown that the large DNA-PKcs molecule forms a distinct structure at the DNA termini, which is likely to play an active role in the formation of a synaptic complex that holds the two ends of the broken DNA molecule together (62,63) (Figure 3B). Small angle X-ray scattering (SAXS) analysis shows the Ku80 C-terminal region may play a role in retaining DNA-PKcs at DSB ends and keeping the DNA-PK complex in a synaptic complex at the DSB site (64).
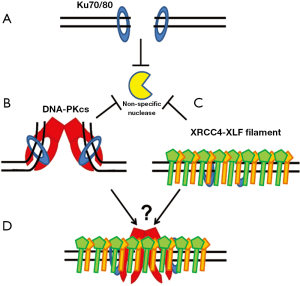
Recently, it has been shown that the XRCC4-XLF complex forms a filament and this filament may also play a role in bridging DNA ends. It was identified that XRCC4 via Arg54, Leu65, and Leu115 in XRCC4 directly interact with the globular head domain of XLF, creating a head to head interface between the two proteins (65). Structural analysis showed that XRCC4 and XLF form super helical filaments via alternating XRCC4 and XLF head domain interfaces which are able to interact with DNA and bridge broken DNA strands (66-68) (Figure 3C). The DNA bridging activity of the XRCC4-XLF filament is consistent with its potential role in stabilizing the DNA ends. The data suggests there may be tethering of DSB ends via Ku-DNA-PKcs and possibly the XRCC4-XLF filament (Figure 3D). A few questions instantly come to mind, including how such a decision is made between DNA-PKcs and XRCC4-XLF to stabilize ends, can DNA-PKcs and the XRCC4-XLF filament bind to the same DSB, and is the decision between the two dependent on the complexity of the break.
DNA end processing
The next step required, if necessary, is processing of the DNA ends to create ligatable ends. Depending on the nature of the break, different DNA end processing enzymes may be required, including those that resect DNA ends, fill in gaps, remove blocking end groups, and make the ends ligatable. There is now a relatively long list of enzymes which have been implicated in being important for processing DNA ends for the NHEJ pathway, including Artemis, PNKP, APLF, Polymerases µ and λ, Werner (WRN), aprataxin, and Ku. As stated above, most of the DNA end processing enzymes are recruited to the DSBs likely by a Ku-XRCC4 scaffold (Figure 2A). A number of factors have been shown to remove blocking end groups in order to make the ends of DSBs ligatable. First, DSB ends can contain non-ligatable 5' hydroxyls or 3' phosphates. PNKP is both a kinase and a phosphatase with the kinase domain responsible for adding a phosphate to a 5' OH and a phosphatase domain for removing 3' phosphate groups (69). Apratxin is a member of the histidine triad family of nucleotide hydrolases and transferases which catalyzes the removal of adenylate groups covalently linked to 5' phosphate termini (70). A surprising new discovery was that Ku has enzymatic activity. Ku was shown to have 5'deoxyribose-5-phosphate (5'-dRP)/AP lyase activity (71). Ku excises abasic sites near DSBs in vitro and this activity was highest when the abasic site was within a short 5' overhang at the DSB end.
The proteins implicated in resecting DNA ends for NHEJ include Artemis, WRN, and APLF. Artemis has been shown to have a number of nucleolytic activities, including a 5' endonucleases activity with a preference to nick a 5' overhang which leaves a blunt duplex end, a 5' to 3' exonuclease activity on single-strand DNA, and the ability to remove 3'-phosphoglycolate groups from DNA termini (72,73). Artemis activity in NHEJ may require the binding to and phosphorylation by DNA-PKcs and/or ATM (Figure 2B). DNA-PKcs mediated-phosphorylation of Artemis is required for its endonucleolytic activity (72). As stated above, WRN interacts with both the Ku heterodimer and XRCC4 and both proteins stimulate the 3' to 5' exonuclease but not 3' to 5' helicase of WRN (45,47,74). APLF is an endonuclease and a 3' to 5' exonuclease (19). None of the other core NHEJ factors modulate APLF nuclease activity on ssDNA or DNA overhangs, however, in reconstituted end joining assays APLF resects 3' overhangs to permit ligation of DNA substrates by XRCC4-DNA Ligase IV (75).
When necessitated by the complexity of the breaks being processed and ligated, filling of gaps of DNA are performed by the family X polymerases which include DNA polymerases µ and λ. Polymerase µ and λ each contain C-terminal BRCT domains which are required for their function (76). Polymerase µ can perform template-dependent synthesis with dNTP and rNTP. When in the presence of Ku and XRCC4/DNA LigaseIV, Pol µ can polymerize across a discontinuous template strand (77). Polymerase λ has template-independent activity and its lyase activity is functional which allows it to remove a damaged base (78).
Ligation of the broken ends and dissolution of the NHEJ complex
The final step in the repair of a DSB is ligation of the broken ends by DNA LigaseIV (Figure 4). DNA ligase IV has activity on its own, but XRCC4 stabilizes LigaseIV which stimulates the ligation activity of LigaseIV (79). Furthermore, XRCC4 stimulates the activity of DNA Ligase IV by promoting the adenylation of DNA Ligase IV (79). DNA LigaseIV can ligate incompatiable DNA ends and ligate across gaps (80). XLF stimulates the activity of activity of DNA Ligase IV towards mismatched and noncohesive DNA ends (81-83). Recently, it was shown that APLF stimulates ligation by XRCC4-DNA LigaseIV above that observed in the presence of only Ku70/80 (18).
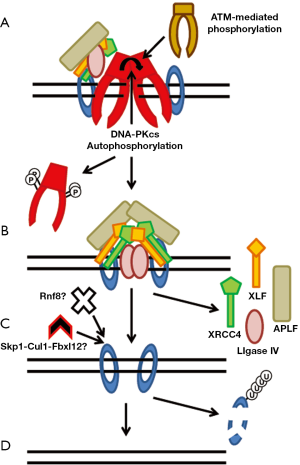
There is not a lot known about the mechanisms which regulate the dissolution of the NHEJ complex from the DSB ends. But, in the past few years a number of studies have looked at the mechanisms required for the ability of NHEJ proteins to dissociate from DNA damage which is believed to occur following completion of repair. Studies with Xenopus egg extracts showed that Ku80 is polyubiquitylated via K48-linkage by Skp1-Cul1-Fbxl12 which mediate the dissociation of Ku from DNA and subsequently results in degradation of Ku by the proteasome (84,85) (Figure 4C). Recent data with human cells implicated that the E3 ubiquitin ligase RNF8 may mediate the dissociation of Ku from DNA ends (86). Depletion of RNF8 resulted in prolonged retention of Ku80 at laser-generated DSBs. The phosphorylation status of DNA-PKcs likely mediates its dissociation from DSBs. Inhibiting phosphorylation at serine 2056 together with the threonine 2609 phosphorylation cluster alters the dynamics of DNA-PKcs at DSB sites resulting in a rigid binding of DNA-PKcs to DNA ends in vivo, which interferes with the NHEJ process (7). Finally, a solution structure of DNA-PKcs obtained by small-angle X-ray scattering (SAXS) revealed that autophosphorylation of DNA-PKcs induces a conformational change which likely releases DNA-PKcs from DNA ends (64) (Figure 4A).
The role of DNA-PKcs kinase activity
As stated above, recruitment of DNA-PKcs to the DNA-Ku complex results in translocation of the Ku heterodimer inward on the DNA, likely to allow DNA-PKcs access to the DNA end resulting in activation of the catalytic activity of the enzyme (87). In the absence of Ku70/80 and DNA, DNA-PKcs has no to limited kinase activity making it truly a DNA dependent kinase (88,89). The kinase domain of DNA-PKcs is surrounded by the FAT and FATC domains. Both domains likely play an important role in regulating the kinase activity of DNA-PKcs and low resolution structures showed that binding to the Ku-DNA complex induces a conformational change in each of these domains and enhancement of DNA-PKcs kinase activity (37-38,90). Deletions in the FATC domain of DNA-PKcs, ATM, and ATR results in a significant decrease of kinase activity suggesting that this domain is important for the activity of the entire PIKK family (91-94). The conformational change induced in the FAT and FATC domains upon interacting with the Ku-DNA complex is predicted to result in the alteration of the catalytic groups and/or the ATP binding pocket of DNA-PKcs resulting in full activation of its kinase activity. Two recent studies have also implicated that the N-terminus plays a role in modulating the enzymatic activity of DNA-PKcs (36,95). Deletion of the N-terminal region of DNA-PKcs and N-terminally restraining DNA-PKcs results in spontaneous activation of its kinase activity. The data suggests that the N-terminus keeps DNA-PKcs basal activity low and that perturbation of the N-terminus likely results in a conformational change which opens the ATP binding pocket of the protein resulting in an increase in basal kinase activity.
Although much is known about the mechanism regulating the activation of the kinase activity of DNA-PKcs, it is still not known what is the exact role the enzymatic activity of DNA-PKcs plays in NHEJ. It is known that the kinase activity of DNA-PKcs is essential for NHEJ as inactivation of DNA-PKcs kinase activity via point mutations or small molecule chemical inhibition results in radiosensitivity and a defect in DSB repair (96). It has been shown that DNA-PKcs can phosphorylate a large number of proteins in vitro including the NHEJ factors Ku70/80, XRCC4, DNA Ligase IV, XLF, WRN, and Artemis and other DNA damage response proteins including H2AX and p53 (5,97). However, the in vivo significance of DNA-PKcs-mediated phosphorylation of these substrates is still not clear. The best characterized DNA-PKcs substrate is DNA-PKcs itself. A large number of the phosphorylation sites are clustered in different regions of DNA-PKcs (98-101). Two prominent phosphorylation clusters which have been identified to be phosphorylated and autophosphorylated in response to IR are the threonine 2609 (T2609) (98,102) and serine 2056 (S2056) cluster (103-105). Phosphorylation of S2056 is a bona fide autophosphorylation site (104) whereas phosphorylation of the T2609 cluster can be phosphorylated by DNA-PKcs itself, ATM, and ATR (105,106). Phosphorylation of these two clusters is important for NHEJ as mutation of phosphorylation sites causes increased radiosensitivity and less efficient DSB repair (99,102,104,107). Ablating phosphorylation of the T2609 cluster and serine 2056 results in rigid binding of DNA-PKcs to laser-generated DSBs in vivo suggesting that phosphorylation at these sites mediates the ability of DNA-PKcs to allow processing of DNA ends for NHEJ and/or its ability to dissociate from DSB ends (7). Single angle X-ray scattering of a hyperphosphorylated form of DNA-PKcs shows that phosphorylation of the enzyme results a conformational change of the protein which opens up the pincers of the molecule and thus illustrating how phosphorylation of the protein may allow it to open ends for processing (64). One phosphorylation site in the kinase domain of DNA-PKcs, threonine 3950, has been characterized (99). Ablation of this site by alanine (T3950A) substitution does not affect the kinase activity of DNA-PKcs but the phospho-mimic aspartic acid (T3950D) substitution results in ablation of DNA-PKcs kinase activity suggesting that autophosphorylation in the kinase domain can regulate the enzymatic activity of DNA-PKcs. Although much has been learned about how the phosphorylation status of DNA-PKcs regulates the functionality of the protein, it is still of great importance to uncover the true substrates of DNA-PKcs in vivo and the role of these phosphorylations in NHEJ and other DSB response.
The role of NHEJ in promoting genome stability and cancer incidence
A fundamental feature of cancer is genome instability. The observation that an increase in cancer frequency is observed in mice and humans with germline mutations in genes that encode for proteins responsible for the response to and repair of DSBs underlie the importance of DSB repair proteins for protecting the genome (2). NHEJ and its factors have long been implicated in playing a role in maintaining genome stability (4). Much of the work detailing the role of NHEJ factors in promoting genome stability have been worked out using mice deficient in one of the NHEJ factors. Fibroblasts from mice deficient in Ku70 and Ku80 show a high frequency of chromosome aberrations including DNA breaks and non-reciprocal translocations (108,109). Furthermore, translocations between DSBs on different chromosomes was increased in the absence of Ku70 in mammalian cells (110). Genomic instability in Ku80 deficient mice was significantly increased in the absence of p53 and these double knock-out mice developed pro B-cell lymphomas and died within 3 months of age (111,112). Ku70 deficient mice have a higher incidence in thymic lymphomas (113). Mouse embryo fibroblasts (MEF) from DNA-PKcs null mice display a higher frequency of spontaneous chromosomal aberrations compared to wild-type mice (108). A rapid onset of lymphomas and leukemias was observed in mice with the DNA-PKcs scid mutant background in the absence of p53 (114). In DNA-PKcs mutant mice in which three phosphorylation sites in the threonine 2609 cluster were ablated via substituting the three amino acids with alanine resulted in hematopoietic stem cell failure caused by excessive DNA damage (115). XRCC4 and DNA ligase IV null mice die during embryogenesis via massive neuronal apoptosis (116,117). However, in a p53 null background which blocks apoptosis in XRCC4 and DNA Ligase IV deficient mice, results in the development of pro-B lymphomas (118,119). Finally, mouse cells from Artemis deficient cells show an increase in chromosomal aberrations (120). No spontaneous Ku mutations have been found in humans suggesting that both Ku70 and Ku80 are likely required for viability. However, two human patients with primary defects in the DNA-PKcs gene have been found. First, a patient with radiosensitive T-B- severe combined immunodeficiency was found to have a missense mutation (L3062R) in the DNA-PKcs gene (121). This missense mutation did not affect overall kinase activity or the DNA end binding capability of DNA-PKcs, but it did affect overall end-joining. It was observed that the L3062R mutation resulted in the inability of DNA-PKcs to activate Artemis and possibly affect the ability of Artemis to open hairpins for the completion of V(D)J recombination. Second, a patient with xeroderma pigmentosum (XP) was also found to be radiosensitive due to a splice variant of DNA-PKcs in which exon 31 was deleted (122). Only clonogenic survival and DSB repair assays were performed using cells obtained from the XP patient; therefore, it is unknown how deletion of exon 31 affects DNA-PKcs activity. Furthermore, studies with human clinical samples have shown a correlation between DNA-PKcs activity and genomic instability and cancer incidence. Dicentric chromosomes and an excess in chromosomal fragmentation increased as DNA-PKcs activity decreases and this decrease in DNA-PKcs activity is associated with a risk in breast and cervical cancer (123). Furthermore, a point mutation at threonine 2609 was identified in a breast tumor sample suggesting that phosphorylation at the T2609 cluster may be important for suppressing breast cancer (124). The role of DNA-PKcs in cancer development and treatment was recently reviewed in much greater detail (125). Cells from human patients with inherited mutations in LigaseIV are radiosensitive, impaired in DSB repair, and have significantly elevated numbers of chromosomal breaks after IR treatment further showing its role in maintaining the human genome (126). Also, polymorphisms within the DNA LigaseIV gene have been linked to a predisposition to multiple myeloma (127). Cells from human patients with mutations in XLF are also radiosensitive and display chromosomal aberrations. And human patients with mutations in Artemis have a predisposition to lymphomas (128). Overall, it is clear that the NHEJ pathway plays a significant role in maintain genome stability and protecting the cell from becoming cancerous.
Conclusions
In this review we summarized the interesting new insights into the mechanism of the NHEJ pathway and the proteins which mediate this repair process. Although these new insights have started to reveal mechanistically how the NHEJ factors are assembled and retained at the DSB and allowed to mediate repair, these insights have opened up new questions. New questions include are there subcomplexes and/or subpathways of classical NHEJ and if so, what factors make up these subcomplexes, what type of DNA DSB does each subcomplex repair, and what mechanism(s) regulate the choice between the subcomplexes for the NHEJ-mediated repair. Lastly, a clear requirement for the kinase activity of DNA-PKcs in NHEJ needs to be elucidated.
Acknowledgments
The authors express their gratitude to Benjamin B.P. Chen for offering suggestions on the review. We apologize to all colleagues whose work we did not specifically mention in this review.
Funding: The research was supported by National Institutes of Health grants (CA162804, CA92584, and CA13499) and Cancer Prevention Research Institute of Texas (RP110465).
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research for the series “DNA Damage and Repair”. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2013.04.02). The series “DNA Damage and Repair” was commissioned by the editorial office without any funding or sponsorship. DJC served as the unpaid Guest Editor of the series and serves as the unpaid editorial board member of Translational Cancer Research. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 2001;27:247-54. [PubMed]
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature 2001;411:366-74. [PubMed]
- Malu S, Malshetty V, Francis D, et al. Role of non-homologous end joining in V(D)J recombination. Immunol Res 2012;54:233-46. [PubMed]
- Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 2006;5:1042-8. [PubMed]
- Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res 2008;18:114-24. [PubMed]
- Mari PO, Florea BI, Persengiev SP, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci U S A 2006;103:18597-602. [PubMed]
- Uematsu N, Weterings E, Yano K, et al. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol 2007;177:219-29. [PubMed]
- Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Biol 2004;5:367-78. [PubMed]
- Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci 2000;25:112-4. [PubMed]
- Zhang Z, Zhu L, Lin D, et al. The three-dimensional structure of the C-terminal DNA-binding domain of human Ku70. J Biol Chem 2001;276:38231-6. [PubMed]
- Zhang Z, Hu W, Cano L, et al. Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions. Structure 2004;12:495-502. [PubMed]
- Mimori T, Hardin JA, Steitz JA. Characterization of the DNA-binding protein antigen Ku recognized by autoantibodies from patients with rheumatic disorders. J Biol Chem 1986;261:2274-8. [PubMed]
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 2001;412:607-14. [PubMed]
- Blier PR, Griffith AJ, Craft J, et al. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem 1993;268:7594-601. [PubMed]
- Costantini S, Woodbine L, Andreoli L, et al. Interaction of the Ku heterodimer with the DNA ligase IV/Xrcc4 complex and its regulation by DNA-PK. DNA Repair (Amst) 2007;6:712-22. [PubMed]
- Nick McElhinny SA, Snowden CM, McCarville J, et al. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol Cell Biol 2000;20:2996-3003. [PubMed]
- Yano K, Morotomi-Yano K, Wang SY, et al. Ku recruits XLF to DNA double-strand breaks. EMBO Rep 2008;9:91-6. [PubMed]
- Grundy GJ, Rulten SL, Zeng Z, et al. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J 2013;32:112-25. [PubMed]
- Kanno S, Kuzuoka H, Sasao S, et al. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J 2007;26:2094-103. [PubMed]
- Macrae CJ, McCulloch RD, Ylanko J, et al. APLF (C2orf13) facilitates nonhomologous end-joining and undergoes ATM-dependent hyperphosphorylation following ionizing radiation. DNA Repair (Amst) 2008;7:292-302. [PubMed]
- Mahajan KN, Gangi-Peterson L, Sorscher DH, et al. Association of terminal deoxynucleotidyl transferase with Ku. Proc Natl Acad Sci U S A 1999;96:13926-31. [PubMed]
- Abraham RT. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amst) 2004;3:883-7. [PubMed]
- Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell 2004;118:9-17. [PubMed]
- Perry J, Kleckner N. The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell 2003;112:151-5. [PubMed]
- Gupta S, Meek K. The leucine rich region of DNA-PKcs contributes to its innate DNA affinity. Nucleic Acids Res 2005;33:6972-81. [PubMed]
- Hartley KO, Gell D, Smith GC, et al. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell 1995;82:849-56. [PubMed]
- Lempiäinen H, Halazonetis TD. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J 2009;28:3067-73. [PubMed]
- Williams DR, Lee KJ, Shi J, et al. Cryo-EM structure of the DNA-dependent protein kinase catalytic subunit at subnanometer resolution reveals alpha helices and insight into DNA binding. Structure 2008;16:468-77. [PubMed]
- Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature 2010;463:118-21. [PubMed]
- Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 1993;72:131-42. [PubMed]
- Gell D, Jackson SP. Mapping of protein-protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res 1999;27:3494-502. [PubMed]
- Singleton BK, Torres-Arzayus MI, Rottinghaus ST, et al. The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol Cell Biol 1999;19:3267-77. [PubMed]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 2005;434:605-11. [PubMed]
- Weterings E, Verkaik NS, Keijzers G, et al. The Ku80 carboxy terminus stimulates joining and artemis-mediated processing of DNA ends. Mol Cell Biol 2009;29:1134-42. [PubMed]
- Jin S, Kharbanda S, Mayer B, et al. Binding of Ku and c-Abl at the kinase homology region of DNA-dependent protein kinase catalytic subunit. J Biol Chem 1997;272:24763-6. [PubMed]
- Davis AJ, Lee KJ, Chen DJ. The N-terminal Region of the DNA-dependent Protein Kinase Catalytic Subunit Is Required for Its DNA Double-stranded Break-mediated Activation. J Biol Chem 2013;288:7037-46. [PubMed]
- Rivera-Calzada A, Spagnolo L, Pearl LH, et al. Structural model of full-length human Ku70-Ku80 heterodimer and its recognition of DNA and DNA-PKcs. EMBO Rep 2007;8:56-62. [PubMed]
- Spagnolo L, Rivera-Calzada A, Pearl LH, et al. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell 2006;22:511-9. [PubMed]
- Hsu HL, Yannone SM, Chen DJ. Defining interactions between DNA-PK and ligase IV/XRCC4. DNA Repair (Amst) 2002;1:225-35. [PubMed]
- Yano K, Morotomi-Yano K, Lee KJ, et al. Functional significance of the interaction with Ku in DNA double-strand break recognition of XLF. FEBS Lett 2011;585:841-6. [PubMed]
- Junop MS, Modesti M, Guarne A, et al. Crystal structure of the Xrcc4 DNA repair protein and implications for end joining. EMBO J 2000;19:5962-70. [PubMed]
- Sibanda BL, Critchlow SE, Begun J, et al. Crystal structure of an Xrcc4-DNA ligase IV complex. Nat Struct Biol 2001;8:1015-9. [PubMed]
- Wu PY, Frit P, Meesala S, et al. Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4. Mol Cell Biol 2009;29:3163-72. [PubMed]
- Mahajan KN, Nick McElhinny SA, Mitchell BS, et al. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol Cell Biol 2002;22:5194-202. [PubMed]
- Cooper MP, Machwe A, Orren DK, et al. Ku complex interacts with and stimulates the Werner protein. Genes Dev 2000;14:907-12. [PubMed]
- Karmakar P, Snowden CM, Ramsden DA, et al. Ku heterodimer binds to both ends of the Werner protein and functional interaction occurs at the Werner N-terminus. Nucleic Acids Res 2002;30:3583-91. [PubMed]
- Kusumoto R, Dawut L, Marchetti C, et al. Werner protein cooperates with the XRCC4-DNA ligase IV complex in end-processing. Biochemistry 2008;47:7548-56. [PubMed]
- Koch CA, Agyei R, Galicia S, et al. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J 2004;23:3874-85. [PubMed]
- Zolner AE, Abdou I, Ye R, et al. Phosphorylation of polynucleotide kinase/ phosphatase by DNA-dependent protein kinase and ataxia-telangiectasia mutated regulates its association with sites of DNA damage. Nucleic Acids Res 2011;39:9224-37. [PubMed]
- Mani RS, Yu Y, Fang S, et al. Dual modes of interaction between XRCC4 and polynucleotide kinase/phosphatase: implications for nonhomologous end joining. J Biol Chem 2010;285:37619-29. [PubMed]
- Iles N, Rulten S, El-Khamisy SF, et al. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol Cell Biol 2007;27:3793-803. [PubMed]
- Clements PM, Breslin C, Deeks ED, et al. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair (Amst) 2004;3:1493-502. [PubMed]
- Yano K, Chen DJ. Live cell imaging of XLF and XRCC4 reveals a novel view of protein assembly in the non-homologous end-joining pathway. Cell Cycle 2008;7:1321-5. [PubMed]
- Reynolds P, Anderson JA, Harper JV, et al. The dynamics of Ku70/80 and DNA-PKcs at DSBs induced by ionizing radiation is dependent on the complexity of damage. Nucleic Acids Res 2012;40:10821-31. [PubMed]
- Yano K, Morotomi-Yano K, Adachi N, et al. Molecular mechanism of protein assembly on DNA double-strand breaks in the non-homologous end-joining pathway. J Radiat Res 2009;50:97-108. [PubMed]
- Rulten SL, Fisher AE, Robert I, et al. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol Cell 2011;41:33-45. [PubMed]
- Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem 2008;283:1-5. [PubMed]
- Pang D, Yoo S, Dynan W, et al. Ku proteins join DNA fragments as shown by atomic force microscopy. Cancer Res 1997;57:1412-5. [PubMed]
- Soutoglou E, Dorn JF, Sengupta K, et al. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol 2007;9:675-82. [PubMed]
- Shao Z, Davis AJ, Fattah KR, et al. Persistently bound Ku at DNA ends attenuates DNA end resection and homologous recombination. DNA Repair (Amst) 2012;11:310-6. [PubMed]
- Nagasawa H, Brogan JR, Peng Y, et al. Some unsolved problems and unresolved issues in radiation cytogenetics: a review and new data on roles of homologous recombination and non-homologous end joining. Mutat Res 2010;701:12-22. [PubMed]
- Cary RB, Peterson SR, Wang J, et al. DNA looping by Ku and the DNA-dependent protein kinase. Proc Natl Acad Sci U S A 1997;94:4267-72. [PubMed]
- Weterings E, van Gent DC. The mechanism of non-homologous end-joining: a synopsis of synapsis. DNA Repair (Amst) 2004;3:1425-35. [PubMed]
- Hammel M, Yu Y, Mahaney BL, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem 2010;285:1414-23. [PubMed]
- Malivert L, Ropars V, Nunez M, et al. Delineation of the Xrcc4-interacting region in the globular head domain of cernunnos/XLF. J Biol Chem 2010;285:26475-83. [PubMed]
- Hammel M, Yu Y, Fang S, et al. XLF regulates filament architecture of the XRCC4.ligase IV complex. Structure 2010;18:1431-42. [PubMed]
- Hammel M, Rey M, Yu Y, et al. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. J Biol Chem 2011;286:32638-50. [PubMed]
- Andres SN, Vergnes A, Ristic D, et al. A human XRCC4-XLF complex bridges DNA. Nucleic Acids Res 2012;40:1868-78. [PubMed]
- Bernstein NK, Williams RS, Rakovszky ML, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol Cell 2005;17:657-70. [PubMed]
- Ahel I, Rass U, El-Khamisy SF, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature 2006;443:713-6. [PubMed]
- Roberts SA, Strande N, Burkhalter MD, et al. Ku is a 5'-dRP/AP lyase that excises nucleotide damage near broken ends. Nature 2010;464:1214-7. [PubMed]
- Ma Y, Pannicke U, Schwarz K, et al. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 2002;108:781-94. [PubMed]
- Povirk LF, Zhou T, Zhou R, et al. Processing of 3'-phosphoglycolate-terminated DNA double strand breaks by Artemis nuclease. J Biol Chem 2007;282:3547-58. [PubMed]
- Perry JJ, Yannone SM, Holden LG, et al. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nat Struct Mol Biol 2006;13:414-22. [PubMed]
- Li S, Kanno S, Watanabe R, et al. Polynucleotide kinase and aprataxin-like forkhead-associated protein (PALF) acts as both a single-stranded DNA endonuclease and a single-stranded DNA 3' exonuclease and can participate in DNA end joining in a biochemical system. J Biol Chem 2011;286:36368-77. [PubMed]
- Moon AF, Garcia-Diaz M, Batra VK, et al. The X family portrait: structural insights into biological functions of X family polymerases. DNA Repair (Amst) 2007;6:1709-25. [PubMed]
- Nick McElhinny SA, Havener JM, Garcia-Diaz M, et al. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell 2005;19:357-66. [PubMed]
- Ramadan K, Shevelev IV, Maga G, et al. De novo DNA synthesis by human DNA polymerase lambda, DNA polymerase mu and terminal deoxyribonucleotidyl transferase. J Mol Biol 2004;339:395-404. [PubMed]
- Grawunder U, Wilm M, Wu X, et al. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 1997;388:492-5. [PubMed]
- Gu J, Lu H, Tippin B, et al. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J 2007;26:1010-23. [PubMed]
- Lu H, Pannicke U, Schwarz K, et al. Length-dependent binding of human XLF to DNA and stimulation of XRCC4.DNA ligase IV activity. J Biol Chem 2007;282:11155-62. [PubMed]
- Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc Natl Acad Sci U S A 2007;104:7851-6. [PubMed]
- Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 2006;124:301-13. [PubMed]
- Postow L, Ghenoiu C, Woo EM, et al. Ku80 removal from DNA through double strand break-induced ubiquitylation. J Cell Biol 2008;182:467-79. [PubMed]
- Postow L, Funabiki H. An SCF complex containing Fbxl12 mediates DNA damage-induced Ku80 ubiquitylation. Cell Cycle 2013;12:587-95. [PubMed]
- Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol 2012;19:201-6. [PubMed]
- Yoo S, Dynan WS. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res 1999;27:4679-86. [PubMed]
- Hammarsten O, Chu G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc Natl Acad Sci U S A 1998;95:525-30. [PubMed]
- West RB, Yaneva M, Lieber MR. Productive and nonproductive complexes of Ku and DNA-dependent protein kinase at DNA termini. Mol Cell Biol 1998;18:5908-20. [PubMed]
- Rivera-Calzada A, Maman JD, Spagnolo L, et al. Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). Structure 2005;13:243-55. [PubMed]
- Banin S, Moyal L, Shieh S, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 1998;281:1674-7. [PubMed]
- Peterson RT, Beal PA, Comb MJ, et al. FKBP12-rapamycin-associated protein (FRAP) autophosphorylates at serine 2481 under translationally repressive conditions. J Biol Chem 2000;275:7416-23. [PubMed]
- Sun Y, Jiang X, Chen S, et al. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A 2005;102:13182-7. [PubMed]
- Beamish HJ, Jessberger R, Riballo E, et al. The C-terminal conserved domain of DNA-PKcs, missing in the SCID mouse, is required for kinase activity. Nucleic Acids Res 2000;28:1506-13. [PubMed]
- Meek K, Lees-Miller SP, Modesti M. N-terminal constraint activates the catalytic subunit of the DNA-dependent protein kinase in the absence of DNA or Ku. Nucleic Acids Res 2012;40:2964-73. [PubMed]
- Kurimasa A, Kumano S, Boubnov NV, et al. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol Cell Biol 1999;19:3877-84. [PubMed]
- Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J 2009;417:639-50. [PubMed]
- Merkle D, Douglas P, Moorhead GB, et al. The DNA-dependent protein kinase interacts with DNA to form a protein-DNA complex that is disrupted by phosphorylation. Biochemistry 2002;41:12706-14. [PubMed]
- Douglas P, Cui X, Block WD, et al. The DNA-dependent protein kinase catalytic subunit is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol Cell Biol 2007;27:1581-91. [PubMed]
- Olsen JV, Vermeulen M, Santamaria A, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 2010;3:ra3. [PubMed]
- Ma Y, Pannicke U, Lu H, et al. The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human Artemis. J Biol Chem 2005;280:33839-46. [PubMed]
- Chan DW, Chen BP, Prithivirajsingh S, et al. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev 2002;16:2333-8. [PubMed]
- Cui X, Yu Y, Gupta S, et al. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol Cell Biol 2005;25:10842-52. [PubMed]
- Chen BP, Chan DW, Kobayashi J, et al. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem 2005;280:14709-15. [PubMed]
- Chen BP, Uematsu N, Kobayashi J, et al. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J Biol Chem 2007;282:6582-7. [PubMed]
- Yajima H, Lee KJ, Chen BP. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol Cell Biol 2006;26:7520-8. [PubMed]
- Ding Q, Reddy YV, Wang W, et al. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol 2003;23:5836-48. [PubMed]
- Ferguson DO, Sekiguchi JM, Chang S, et al. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc Natl Acad Sci U S A 2000;97:6630-3. [PubMed]
- Karanjawala ZE, Grawunder U, Hsieh CL, et al. The nonhomologous DNA end joining pathway is important for chromosome stability in primary fibroblasts. Curr Biol 1999;9:1501-4. [PubMed]
- Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol 2007;9:978-81. [PubMed]
- Nussenzweig A, Sokol K, Burgman P, et al. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival, and development. Proc Natl Acad Sci U S A 1997;94:13588-93. [PubMed]
- Difilippantonio MJ, Zhu J, Chen HT, et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 2000;404:510-4. [PubMed]
- Gu Y, Seidi K, Rathbun G, et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 1997;7:653-65. [PubMed]
- Guidos CJ, Williams CJ, Grandal I, et al. V(D)J recombination activates a p53-dependent DNA damage checkpoint in scid lymphocyte precursors. Genes Dev 1996;10:2038-54. [PubMed]
- Zhang S, Yajima H, Huynh H, et al. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J Cell Biol 2011;193:295-305. [PubMed]
- Gao Y, Sun Y, Frank KM, et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell 1998;95:891-902. [PubMed]
- Barnes DE, Stamp G, Rosewell I, et al. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr Biol 1998;8:1395-8. [PubMed]
- Gao Y, Ferguson DO, Xie W, et al. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature 2000;404:897-900. [PubMed]
- Frank KM, Sharpless NE, Gao Y, et al. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol Cell 2000;5:993-1002. [PubMed]
- Rooney S, Sekiguchi J, Zhu C, et al. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol Cell 2002;10:1379-90. [PubMed]
- van der Burg M, Ijspeert H, Verkaik NS, et al. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest 2009;119:91-8. [PubMed]
- Abbaszadeh F, Clingen PH, Arlett CF, et al. A novel splice variant of the DNA-PKcs gene is associated with clinical and cellular radiosensitivity in a patient with xeroderma pigmentosum. J Med Genet 2010;47:176-81. [PubMed]
- Someya M, Sakata K, Matsumoto Y, et al. The association of DNA-dependent protein kinase activity with chromosomal instability and risk of cancer. Carcinogenesis 2006;27:117-22. [PubMed]
- Wang X, Szabo C, Qian C, et al. Mutational analysis of thirty-two double-strand DNA break repair genes in breast and pancreatic cancers. Cancer Res 2008;68:971-5. [PubMed]
- Hsu FM, Zhang S, Chen BP. Role of DNA-dependent protein kinase catalytic subunit in cancer development and treatment. Transl Cancer Res 2012;1:22-34. [PubMed]
- Riballo E, Critchlow SE, Teo SH, et al. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr Biol 1999;9:699-702. [PubMed]
- Roddam PL, Rollinson S, O’Driscoll M, et al. Genetic variants of NHEJ DNA ligase IV can affect the risk of developing multiple myeloma, a tumour characterised by aberrant class switch recombination. J Med Genet 2002;39:900-5. [PubMed]
- Moshous D, Pannetier C. Partial T and B lymphocyte immunodeficiency and predisposition to lymphoma in patients with hypomorphic mutations in Artemis. J Clin Invest 2003;111:381-7. [PubMed]


