
Alternative pathways of non-homologous end joining (NHEJ) in genomic instability and cancer
Background on DSB repair
DNA double-strand breaks (DSB) are generated in a random manner in the genome by exogenous agents, such as ionizing radiation (IR) or radiomimetic drugs. Endogenous events leading to accidental DSBs also include oxidative damage, replication fork collapse and telomere erosion. DSBs also occur as programmed events during meiosis, as well as during V(D)J recombination and class switch recombination (CSR) required for immunoglobulin diversity and function. Accidental or programmed DSBs, if left unrepaired or if repaired in an erroneous manner, can have severe adverse consequences for the genome including the generation of mutations and chromosomal aberrations. Both forms of genomic alterations are implicated in cell death (1,2), as well as in genomic instability leading to the development of cancer (3).
To maintain genomic integrity, cells have evolved several pathways to process DSBs and mitigate their adverse consequences. The two key DSB repair pathways engaged to this task are non-homologous end joining (NHEJ) and homologous recombination repair (HRR) (4-7).
The classical or canonical form of NHEJ is a very fast process operating with half times of 10-30 min. It functions by simply joining the DNA ends and has no build-in potential of restoring the original sequence in the vicinity of the DSB (Figure 1). Essential components of NHEJ are the three subunits of the DNA-PK complex (Ku70, Ku80, DNA-PKcs), and the LIG4/XRCC4/XLF complex (8-12). As this pathway relies on the evolutionarily new DNA-PKcs, it is sometimes termed as D-NHEJ. However, the term classical or canonical NHEJ (C-NHEJ) is more frequently used and is also adopted here [reviewed in (7,13,14)].
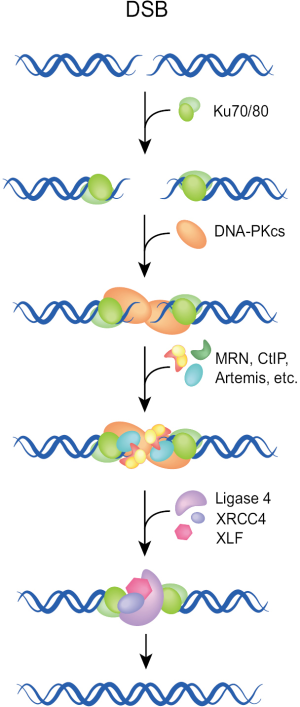
C-NHEJ is error-prone on two counts: first, it has no build-in mechanisms ensuring the restoration of the original DNA sequence in the vicinity of the DSB. As a result, it is associated with sequence alterations including random exchanges of nucleotides, as well as the addition or deletion of several base pairs at the junction. These events are more likely to occur during repair of radiation-induced DSBs, due to the end-processing requirement for the generation of ligatable DNA ends. Second, C-NHEJ has no build-in mechanisms ensuring the restoration of the original DNA molecule and can in principle join any DNA ends irrespective of molecular origin. As a result, the generation of new sequence-combinations is possible, which in higher eukaryotes can manifest as chromosomal translocations. Possibly as a direct consequence of its operational speed, C-NHEJ is associated with more limited sequence alterations at the junction and has a lower probability of translocation formation than end-joining pathways operating with slower kinetics (see below). As a result, C-NHEJ is considered a guardian of genomic stability and suppressor of carcinogenesis (15-17). The molecular determinants underpinning the high speed of C-NHEJ remain uncharacterized.
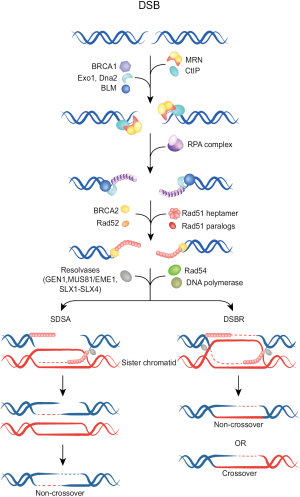
In contrast to C-NHEJ, HRR operates with slower kinetics and requires a homologous template to not only repair the DSB, but to also restore the sequence around the break. Higher eukaryotes use the sister chromatid as a homologous template, and as a direct consequence HRR is restricted to the S and G2 phases of the cell cycle. Initial step in HRR is DNA nucleolytic end resection by the MRN complex (comprising Mre11, Rad50, Nbs1) along with other accessory proteins such as CtIP and the tumor suppressor protein BRCA1. An eminent role for the long-range resection have the Bloom helicase (BLM), Exonuclease 1 (Exo1) and Dna2 helicase/nuclease (Figure 2) (23-25). Thereby, terminal nucleotides in the 5' ends are removed generating long 3' single-stranded DNA (ssDNA) overhangs on both sides of the break. These 3'-ssDNA tails, representing the substrate for HR repair machinery, are coated and stabilized by the Replication protein A (RPA) complex, which subsequently becomes displaced by Rad51 recombinase generating Rad51 nucleoprotein filament. After homology search, strand invasion with the donor DNA and D-loop formation, a polymerase catalyzes DNA synthesis until finally the Holliday junctions become resolved, resulting in a crossover or non-crossover product (for a detailed description of this process and the participating proteins see in Figure 2) (22).
In addition to C-NHEJ and HRR, recent studies demonstrate the operation of a third pathway of DSB processing, functioning on simple end-joining principles, but repairing DSBs slower (t50 30 min to 20 h) than C-NHEJ (9,16,26-28). This repair pathway is considered to be an alternative form of NHEJ and is frequently abbreviated as A-NHEJ, or simply A-EJ (29). Since A-EJ is suppressed by C-NHEJ, and possibly also by HRR, and gains functional relevance when these standard repair processes fail, globally or locally, it is also considered to be a backup pathway and has been abbreviated as B-NHEJ (6,7). Throughout this review we will use the term A-EJ to refer to this repair pathway.
Although A-EJ does not require homology for function, as for example HRR does, it is occasionally facilitated by microhomologies fortuitously found at the DNA ends, particularly when resection and the generation of single stranded DNA regions precedes end-joining. The form of DSB processing utilizing microhomologies is frequently termed microhomology-mediated end-joining (MMEJ). The occasional use of microhomologies in this repair pathway makes A-EJ a term semantically preferable to A-NHEJ. In the following discussion we consider MMEJ as a subset of A-EJ and refer explicitly to it only if it is relevant to the discussed context [reviewed in (7,29-32)].
A-EJ, as C-NHEJ, is error prone on two counts: it has no build-in mechanisms for restoring the DNA sequence in the vicinity of the DSB, and can catalyze the joining of unrelated DNA molecules, leading thus to the formation of translocations (7,16,32-34). On both counts, A-EJ is more error prone than C-NHEJ, i.e., sequence alterations at the junctions are more frequent and more extensive and the probability of translocation formation is much higher. As a result, and in contrast to C-NHEJ that is considered as a guardian of genomic stability, A-EJ is considered a major source of genomic instability (17,35-37).
The above outlined basic features of C-NHEJ, A-EJ and HRR indicate distinct functional characteristics, distinct possible outcomes during normal operation, and widely different risks for errors. These large and fundamental differences make the question of repair pathway choice for the processing of each individual DSB difficult to answer. Correct repair is only afforded by HRR, but is only available in a fraction of the cell cycle. C-NHEJ functions throughout the cell cycle and removes DSBs with high efficiency at the cost of small sequence alterations and a low probability for translocations. Finally, A-EJ functions throughout the cell cycle but is clearly functionally enhanced in S and G2 (38-42); it has the highest probability for generating translocations, as well as large deletions and other sequence alterations at the junction. Such dramatic differences in features, outcomes and risks make simple competition an “unwise” mechanism for pathway selection and suggest that undefined parameters underpin repair pathway choice. Such candidate parameters are presently hotly debated, but the issue remains for the most part unresolved.
In the following sections, we review the known enzymatic requirements of A-EJ and discuss emerging evidence for the involvement of this pathway in the generation of genomic instability and the development of cancer.
Core components of A-EJ
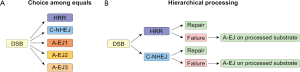
Several factors have been implicated in A-EJ (Figure 3) and their functional diversity has led to the postulate that there are several sub-pathways in operation, engaging prospectively at each DSB on the basis of as of yet undefined parameters in competition with other repair pathways (Figure 4A). A different model emerges by regarding all functions of A-EJ as backup operations that are initiated at DSBs only after C-NHEJ or HRR have engaged but failed to successfully complete processing (Figure 4B). According to the former model, the recruitment at the ends of factors of a given repair pathway will determine the processing of the DSB by this pathway. However, according to the second model, A-EJ will engage at DSBs where either C-NHEJ or HRR have attempted processing but somehow failed. Thus, at each DSB where A-EJ engages, factors of either C-NHEJ or HRR, particularly those involved at early steps, will be present when A-EJ takes DSB processing over. Also, it is possible, and even likely, that these factors have already operated at DNA ends and have carried out one or more of the initial steps of C-NHEJ or HRR, which of course alters the state of the substrate presented to A-EJ. Furthermore, the presence of C-NHEJ and HRR factors at the DNA ends may either facilitate or compromise A-EJ. In the following paragraphs, we review the function of A-EJ and the factors implicated from this mechanistic/operational perspective.
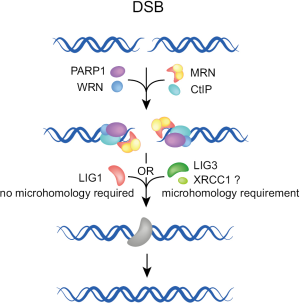
When the engagement of A-EJ follows failure of C-NHEJ, several of the early C-NHEJ factors may be present at the junction, but the process must be abrogated prior to ligation by LIG4. Available evidence suggests that under these conditions, end ligation is possible with one of the remaining ligases, LIG3 and LIG1 (43-48). While LIG3 is likely to be more effective, LIG1 is also remarkably fit for this function (48). Since LIG3 operates in other repair pathways together with XRCC1 and PARP1, its involvement in A-EJ implicates these factors in A-EJ as well. PARP1 is a sensor for DNA discontinuities, originally shown to operate in base excision and single-strand break repair (49). Previous work implicated PARP1 also in repair by A-EJ (46,50-53). There is even evidence for competition between Ku and PARP1 for DSBs (50,54,55) raising the possibility that pre-existing C-NHEJ factors at the DSB compromise A-EJ.
DNA end stabilization provided in C-NHEJ by Ku may be provided in A-EJ by histone H1 (56). However, it should be emphasized that to date the evidence for a role of histone H1 in A-EJ is of purely biochemical nature. Backup of C-NHEJ by A-EJ is likely to be associated with increased processing at the ends, and to result in the more frequent use of microhomologies. This form of backup function can occur in all phases of the cell cycle, because C-NHEJ is active throughout the cell cycle.
However, it is also possible that A-EJ backs up failures of HRR. Such events are possible only when HRR is active, i.e., in the S- and G2-phase of the cell cycle. It will also pertain only for the subset of DSBs that are processed by this repair pathway, and which according to current estimates lies between 10-20% of radiation induced DSBs (57,58). Under these conditions A-EJ may operate on resected ends that must be generated in order to inactivate C-NHEJ and allow end resection to prepare for HRR. This may explain the observed dependence of A-EJ on the MRN complex as well as CtIP, BRCA1 etc (45,59-65). Also, as a result of the processed ends likely to be present, it is possible that microhomologies fortuitously present are used as means of intermolecular stabilization of the two DNA ends to efficiently complete rejoining (29). However, end joining using microhomologies was also reported in vitro in reactions setup with cell-free extracts (66), and has also been described between repetitive elements in the human genome through single-strand annealing leading to genome rearrangements (67).
A-EJ is considered to be a mechanistically distinct repair pathway, and has been shown to be active throughout the cell cycle (7,16,31,33,68). Notably though, it is markedly enhanced in the G2 as compared to G1 phase, and is compromised in stationary-phase cells tested either in the G1 or G2 phase of the cell cycle (31,41,42,69). There are speculations that the latter response may be regulated by phosphorylation of BRCA1 at S988 through Chk2, where in its phosphorylated form BRCA1 promotes error-free NHEJ and suppresses mutagenic A-EJ. Therewith, it reduces the size of deletions at the breakpoint junction (65,70-72). However, this dependency is more likely in G2 than in G1 cells as BRCA1/CtIP/MRN initiates DSB resection during S/G2 phases (73), and therefore alternative mechanisms should be explored.
A-EJ and the development of leukemias
Chromosomal translocations are a type of rearrangements, where parts of two different non-homologous chromosomes fuse together. Such types of chromosomal abnormalities have been described in cancer, mainly in leukemia and many types of lymphoma (34,74-77). Several lines of evidence implicate A-EJ to chromosome translocation formation that underlie leukemia and lymphoma (36,37,78,79). Since these cancers frequently originate from erroneous repair of programmed DSBs generated during V(D)J and CSR recombination, we briefly review these processes before summarizing the available evidence for role of A-EJ in their formation.
V(D)J recombination, also known as somatic recombination, and class switch recombination (CSR) are fundamental physiological mechanisms for the generation of the immune repertoire in mammals and for the development and survival of B and T cells. In both processes DNA DSBs occur as programmed events and are necessary for triggering recombination.
V(D)J recombination aims the generation of different combinations of pre-existing gene segments: variable (V), diversity (D) and joining (J). The first step is cleavage within specific recombination signal sequences (RSSs) by the lymphocyte-specific endonucleases RAG1 and RAG2, operating as a complex called RAG. These segments are rearranged next to each other during the resolution of these DSBs that are rejoined almost exclusively by C-NHEJ resulting in a V(D)J segment [for a review see (80-85)].
CSR is a process occurring in antigen-stimulated mature B cells, which changes the antibody production from one immunoglobulin (Ig) class or subclass (isotype) to another—from IgM to IgG, IgE or IgA. The constant regions of different Ig isotypes are encoded by distinct CH exon clusters in the immunoglobulin heavy chain locus. CSR is initiated by activation-induced cytidine deaminase (AID), which deaminates cytidines to uridines within switch (S) regions. The ensuing cascade of repair reactions leads to the generation of DSBs. Deleting sequences between the S regions leads to expression of a new constant region, and thus to the production of an antibody of different class. Hence, during CSR the heavy chains are altered but the variable regions remain unchanged and the antibody retains its antigen specificity [for a detailed review see (86,87)].
Several NHEJ factors have already been implicated in V(D)J recombination (88-92). However, evidence exists that the essential component of C-NHEJ, Ku70, is dispensable for T cell antigen receptor (TCR) V(D)J recombination, suggesting the involvement of other repair pathways (93).
Deficiencies in the immune system development, particularly lymphocytes, lead to severe combined immunodeficiency syndrome (SCID), which is often associated with mutations in DNA repair proteins (94-99). Notably, the SCID phenotype is observed in mice deficient in any of C-NHEJ proteins (100,101), as well as in mice lacking one of the RAG proteins (81,102,103). These mice also frequently develop tumors with translocations involving the Ig locus that are now known to be generated by A-EJ (see below). A model was proposed, where RAG1/2 proteins together with C-NHEJ factors suppress A-EJ mediated genomic instability during V(D)J recombination (104,105). This result could also be reproduced in a plasmid model system (106).
It is relevant to point out here that RAG post-cleavage complex shunts the broken DNA ends to C-NHEJ, thus suppressing aberrant recombination events. In compromised V(D)J recombination, through mutations destabilizing the post-cleavage complex, the ends are free to participate in HRR or A-EJ repair leading to CSR but also to genomic instability (104,105,107-109).
Although RAG-mediated DSBs during V(D)J recombination are predominantly rejoined by the C-NHEJ pathway, repair of DSBs in the switch (S) regions during CSR is not largely compromised in C-NHEJ deficient cells and a direct shift of processing to A-EJ is observed (110-112). CSR is not affected by the absence of DNA-PKcs (113,114), and is likely to use Lig1 or Lig3 (44). Taking into account the requirement of Ku for CSR (115,116), these findings strongly suggest the use of A-EJ during CSR.
As mentioned above, A-EJ frequently joins IgH locus breaks to breaks in genes such as c-myc in other chromosomes generating translocations and causing leukemias. This has been observed in the combined absence of Ku70 and Lig4, as well as in the absence of XRCC4 (36,110). Besides normal CSR frequency, B cells heterozygous for XRCC1 also show reduced IgH/c-myc translocations during CSR implicating XRCC1 in A-EJ (117). An interesting observation is that this proposed factor for A-EJ, XRCC1, which acts in a complex with Lig3, is not required for A-EJ during CSR and its absence even slightly increases CSR efficiency (118,119). Similar results were also obtained with mouse cells deleted of different forms of LIG3 (120). Also PARP1 and PARP2 do not seem to be required for CSR. However, PARP1 was shown to favor A-EJ and PARP2 to suppress translocations during CSR (121). Interestingly, in I.29µ B cell lymphoma and splenic B cells, PARP inhibitors lead to increased antibody class switching (122). It appears therefore that the context of the DSB determine the requirements for components such as PARP1 and XRCC1. In this regard, DSB repair by PARP1-dependent/Ku-independent EJ is more efficient in the presence of microhomology termini containing G:C base pairs (123,124).
An A-EJ pathway using microhomologies has been invoked in a general model of formation of oncogenic complex translocations (complicons). Pro-B lymphomas in mice lacking both p53 and classical NHEJ contain complicons that co-amplify c-myc sequence in chromosome 15 and IgH in chromosome 12 associated with microhomology at the translocation junctions (125). Furthermore, deficiency of C-NHEJ component XLF/Cernunnos is also associated with human B cell malignancies, where CSR junctions are also characterized by long microhomologies (126).
The expression of oncogenic BCR-ABL gene fusion is a result of reciprocal translocation t[9;22] and is predominantly associated with chronic myelogenous leukemia (CML). BCR-ABL tyrosine kinase facilitates cell division and results in increased reactive oxygen species (ROS), which in turn lead to increased DNA damage including DSBs (127,128). Moreover, BCR-ABL-positive CML is associated with up-regulated A-EJ (129).
The BCR-ABL translocation is often associated with microhomologies at the junctions and with interspersed repeats (130). Furthermore, in BCR-ABL-positive CML cells key proteins of C-NHEJ, Artemis and Lig4, are down-regulated. In contrast, the levels of proteins involved in A-EJ, Lig3α and WRN, are elevated. Additionally, depletion of either Lig3α or WRN results in decreased end-joining efficiency. The authors suggest therefore that A-EJ enables CML cells to repair ROS-induced DSBs and survive. Since A-EJ is error-prone, the survival is associated with increased genomic instability and disease progression (131).
The most common mutations in acute myeloid leukemia (AML) are internal tandem duplications (ITD) of FMS––like tyrosine kinase-3 (FLT3) receptor, known as FLT3-ITD. Cells expressing FLT3-ITD and bone marrow mononuclear cells from FLT3-ITD knock-in mice utilize microhomology-mediated A-EJ to repair DSBs leading to increased number of deletions. Additionally, the level of Lig3α in FLT3-ITD-expressing cells is up-regulated and the protein level of the C-NHEJ component Ku is decreased, indicating that the FLT3 signaling pathway shifts DSB repair toward A-EJ (132).
But how is C-NHEJ suppressing chromosome translocations? In a model for DSB rejoining of correct ends, DNA-PKcs has a key role: together with Ku, DNA-PKcs improves the interactions between the participating proteins generating thus some form of molecular rejoining machine (16,133,134). Additionally, the local chromatin structure is altered and the correct ends are rapidly captured within this machine. In contrast, the evolutionarily older and slower A-EJ is using proteins that fail to undergo efficient intermolecular interactions, reducing thus efficiency and speed of repair and increasing the probability of end resection and exchange formation (16).
This model is supported by the observation that the C-NHEJ protein Ku80 ensures positional stability of broken DNA ends (135) and suppresses chromosomal aberrations in mouse cells (136). Along this line, cells deficient in Ku70 display increase in reciprocal translocations (137).
Moreover, a detailed work by Jasin et al. implicates A-EJ as a main pathway for chromosomal translocations in mammalian cells. The C-NHEJ component XRCC4-Lig4 suppresses A-EJ-mediated translocation formation in wild-type cells. The authors found longer microhomologies at the junctions in XRCC4-/- cells, and that DSB repair causing translocations does not rely on XRCC4 (37). These observations are further supported by the requirement of Lig3 for microhomology-mediated EJ during translocation formation (44). Finally, there are speculations about a second alternative of EJ, which acts independently of microhomologies and utilizes Lig1 (44). An interesting observation is that the high level of genomic instability of bladder cancer is a consequence of microhomology-mediated end-joining of DSBs, which is likely a main repair pathway in bladder cancer cells (138).
Thus, A-EJ is more translocation prone, probably due to its mechanistic basis and its slower kinetics, and therewith responsible for genomic instability and cancer development.
The function of A-EJ in telomere maintenance
If not protected, chromosome ends (telomeres) are likely to be recognized as broken DNA ends and to elicit DNA damage response (DDR) (139,140). As a direct consequence of the initiated DDR, different chromosomes can fuse end-to-end by NHEJ and less frequently by homology-mediated repair, resulting in dicentric chromosomes, which are unstable during mitosis. Similar unfavorable effects are initiated when the chromatids of one chromosome join to form a sister union that can cause anaphase bridges leading to genomic instabilities or cell death.
To escape this problem, the ends of eukaryotic chromosomes are protected by telomeres, which also circumvent the sequence loss problem associated with semi-conservative DNA replication. Telomeres are nucleoprotein structures, consisting of long stretches of TTAGGG repeats in humans and the telomere-specific protein complex, shelterin. The key enzyme in telomere maintenance, which adds telomeric repeats to the 3' end of DNA strands, is the telomerase. Moreover, end protection benefits from the unique structure of the telomere, t-loop [for a detailed review, see (141)].
Previous work suggested that NHEJ is the major DNA repair pathway responsible for the fusion of dysfunctional telomeres (142). But which pathway is primarily responsible for this function? A plenty of data shows that C-NHEJ proteins like DNA-PKcs and Ku are present at the telomeric regions and are actually required for telomere capping/maintenance (143-148). The presence of these proteins in the telomeric regions suggests that in case of telomere erosion, end-joining by C-NHEJ may ensue. However, using a telomerase-deficient mouse model, Maser and colleges reported that fusion of critically shortened telomeres does not depend on the C-NHEJ components DNA-PKcs and Lig4, and suggested that A-EJ has a major role (149). Other reports suggest that dysfunctional telomeres may be processed for joining by both NHEJ pathways, although A-EJ may be preferentially functional on naturally shortened telomeres (150).
A recent study from Oh and colleges reports the synthetic lethality between Lig4 and Rad54B in human epithelial cells, suggesting that A-EJ is not sufficient for repair of DSBs. In addition, they postulate that C-NHEJ often leads to chromosome: chromosome fusions, while A-EJ favors sister chromatid fusions (151). A proposed model is that in wild type cells, Ku and the shelterin subunit TRF2 suppress both C-NHEJ and A-EJ. However, overexpression of the dominant-negative TRF2ΔBΔM to suppress TRF2 function leads to increased number of chromosome: chromosome fusions supported by C-NHEJ. On the other hand, A-EJ dominates in Ku86 deficient cells resulting in large numbers of sister: sister chromatid fusions (151). These data are reminiscent of the PARP1-Ku competition and suggest that telomere erosion follows similar rules.
A-EJ factors as targets for cancer therapy
A novel and promising therapeutic strategy is the use of PARP1 inhibitors to improve the therapy of cancers with BRCAness (152). Similarly, PARP1 inhibitors could be implicated in the therapy of cancers associated with increase in the use of A-EJ pathway for DSB repair.
Attractive therapeutic targets are also the DNA ligases, which complete the process by rejoining the DNA ends (153). As already described above, the levels of two proteins involved in A-EJ, Lig3α and Werner syndrome helicase (WRN), are up-regulated in BCR-ABL-positive CML cells (131). Since BCR-ABL-positive CML is treated by the tyrosine-kinase inhibitor Imatinib (Gleevec), the inhibition of A-EJ factors reveals a novel and more effective therapeutic approach. A recent study from Tobin and colleagues reported that BCR-ABL-positive CML cells resistant to Imatinib were hypersensitive to the combined treatment of Ligase and PARP inhibitors, correlating with hyperactive A-EJ. Furthermore, elevated levels of Lig3α and PARP1 in CML patients were proposed to be biomarkers for therapies targeting A-EJ components when treatment with tyrosine kinase inhibitors is ineffective (154). This novel therapeutic approach could also be applied to therapy-resistant breast cancer cell lines, which were shown to be sensitive to DNA ligase and PARP inhibitors (155). Similarly, new therapeutic strategies for AML associated with FLT3 mutations may also include Lig3α or PARP1 inhibitors (132).
Concluding remarks
The involvement of A-EJ in genome instability and cancer development is indisputable. However, the mechanisms balancing C-NHEJ and A-EJ in healthy cells remain unknown and require in-depth study.
Parameters and proteins having positive and negative effects on A-EJ are depicted in Figure 5. As already outlined above, Ku ensures positional stability of broken DNA ends (135) and competes with PARP1 for DSB repair (50,54,55). In addition, Ku is considered the main determinant of the choice between C-NHEJ and A-EJ in human somatic cells, preferring to assist C-NHEJ (156). Another DNA damage response factor, 53BP1, also favors CSR through C-NHEJ by preventing DNA end resection and A-EJ (157,158). In addition, histone variant H2AX suppresses DNA end resection in G1-phase lymphocytes ensuring efficient repair by C-NHEJ (159). Finally, Fanconi anemia (FA) genes contribute to the regulation of NHEJ pathways (160), and there is evidence that DNA-PKcs negatively regulates A-EJ (161). While this information generates solid foundations to build upon, important details are still missing and require further exploration.
Besides proper DNA repair, the maintenance of genome integrity requires checkpoint activation. In this regard, ATM was shown to prevent prolonged presence of DSBs and chromosomal translocations in lymphocytes (162). ATM is also directly involved in RAG-induced DSB repair ensuring stable DSB complexes and preventing aberrant rearrangements (163). Possible connections between checkpoint proteins and A-EJ regulation remain to be established.
In the front of cancer treatment, the possibility of combining inhibitors of A-EJ with other treatment modalities to improve the outcome, at least in tumors with enhanced A-EJ, has strong rationale and is likely to see application in the future. Finally, the intriguing possibility of protecting organisms from carcinogenesis by limiting the function of A-EJ should be considered and tested.
Acknowledgments
Funding: Work supported by grants from the DFG (GRK1739), the “Bundesministerium für Bildung und Forschung” (BMBF: 02NUK005C and 03NUK001B) and the “Bundesministerium für Wirtschaft und Technologie” (BMWi: ESA-AO-08-IBER, 50WB1229).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (David J. Chen and Benjamin P.C. Chen) for the series “DNA Damage and Repair” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2013.05.02). The series “DNA Damage and Repair” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hall EJ, Giaccia AJ. eds. Radiobiology for the Radiologist. Philadelphia, Baltimore, New York, London, Buenos Aires, Hong Kong, Sydney, Tokyo: Lippincott Williams & Wilkins, 2006.
- Friedberg EC, Walker GC, Siede W, et al. eds. Ellenberger, DNA Repair and Mutagenesis. Washington, DC, USA: ASM Press, 2006.
- Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev 2003;194:77-95. [PubMed]
- Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet 2006;40:363-83. [PubMed]
- Helleday T, Lo J, van Gent DC, et al. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923-35. [PubMed]
- Kinner A, Wu W, Staudt C, et al. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res 2008;36:5678-94. [PubMed]
- Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat Res 2011;711:61-72. [PubMed]
- Li Z, Otevrel T, Gao Y, et al. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell 1995;83:1079-89. [PubMed]
- DiBiase SJ, Zeng ZC, Chen R, et al. DNA-dependent protein kinase stimulates an independently active, nonhomologous, end-joining apparatus. Cancer Res 2000;60:1245-53. [PubMed]
- Wang H, Zeng ZC, Perrault AR, et al. Genetic evidence for the involvement of DNA ligase IV in the DNA-PK-dependent pathway of non-homologous end joining in mammalian cells. Nucleic Acids Res 2001;29:1653-60. [PubMed]
- Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 2006;124:301-13. [PubMed]
- Ding Q, Reddy YV, Wang W, et al. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol 2003;23:5836-48. [PubMed]
- Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J 2009;417:639-50. [PubMed]
- Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem 2008;283:1-5. [PubMed]
- Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 2006;5:1042-8. [PubMed]
- Iliakis G, Wang H, Perrault AR, et al. Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenet Genome Res 2004;104:14-20. [PubMed]
- Ferguson DO, Sekiguchi JM, Chang S, et al. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc Natl Acad Sci U S A 2000;97:6630-3. [PubMed]
- Suwaki N, Klare K, Tarsounas M. RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin Cell Dev Biol 2011;22:898-905. [PubMed]
- Ip SC, Rass U, Blanco MG, et al. Identification of Holliday junction resolvases from humans and yeast. Nature 2008;456:357-61. [PubMed]
- Ciccia A, Constantinou A, West SC. Identification and characterization of the human mus81-eme1 endonuclease. J Biol Chem 2003;278:25172-8. [PubMed]
- Rass U, Compton SA, Matos J, et al. Mechanism of Holliday junction resolution by the human GEN1 protein. Genes Dev 2010;24:1559-69. [PubMed]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 2008;77:229-57. [PubMed]
- Nimonkar AV, Ozsoy AZ, Genschel J, et al. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci U S A 2008;105:16906-11. [PubMed]
- Nimonkar AV, Genschel J, Kinoshita E, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 2011;25:350-62. [PubMed]
- Karanja KK, Cox SW, Duxin JP, et al. DNA2 and EXO1 in replication-coupled, homology-directed repair and in the interplay between HDR and the FA/BRCA network. Cell Cycle 2012;11:3983-96. [PubMed]
- Wang H, Perrault AR, Takeda Y, et al. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res 2003;31:5377-88. [PubMed]
- Kabotyanski EB, Gomelsky L, Han JO, et al. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res 1998;26:5333-42. [PubMed]
- Terzoudi GI, Singh SK, Pantelias GE, et al. Premature chromosome condensation reveals DNA-PK independent pathways of chromosome break repair. Int J Oncol 2008;33:871-9. [PubMed]
- McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet 2008;24:529-38. [PubMed]
- Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell 2007;131:223-5. [PubMed]
- Iliakis G. Backup pathways of NHEJ in cells of higher eukaryotes: cell cycle dependence. Radiother Oncol 2009;92:310-5. [PubMed]
- Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol 2012;116:1-49. [PubMed]
- Iliakis G, Wu W, Wang M, et al. Backup pathways of nonhomologous end joining may have a dominant role in the formation of chromosome aberrations. in chromosomal alterations (G. Obe, Vijayalaxmi, Ed.), pp. 67-85. Springer Verlag, Berlin, Heidelberg, New York, 2007.
- Gostissa M, Alt FW, Chiarle R. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu Rev Immunol 2011;29:319-50. [PubMed]
- Lieber MR. NHEJ and its backup pathways in chromosomal translocations. Nat Struct Mol Biol 2010;17:393-5. [PubMed]
- Boboila C, Jankovic M, Yan CT, et al. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci U S A 2010;107:3034-9. [PubMed]
- Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol 2010;17:410-6. [PubMed]
- Wu W, Wang M, Mussfeldt T, et al. Enhanced use of backup pathways of NHEJ in G2 in Chinese hamster mutant cells with defects in the classical pathway of NHEJ. Radiat Res 2008;170:512-20. [PubMed]
- Guirouilh-Barbat J, Huck S, Lopez BS. S-phase progression stimulates both the mutagenic KU-independent pathway and mutagenic processing of KU-dependent intermediates, for nonhomologous end joining. Oncogene 2008;27:1726-36. [PubMed]
- Wu W, Wang M, Wu W, et al. Repair of radiation induced DNA double strand breaks by backup NHEJ is enhanced in G2. DNA Repair (Amst) 2008;7:329-38. [PubMed]
- Singh SK, Bednar T, Zhang L, et al. Inhibition of B-NHEJ in plateau-phase cells is not a direct consequence of suppressed growth factor signaling. Int J Radiat Oncol Biol Phys 2012;84:e237-43. [PubMed]
- Singh SK, Wu W, Zhang L, et al. Widespread dependence of backup NHEJ on growth state: ramifications for the use of DNA-PK inhibitors. Int J Radiat Oncol Biol Phys 2011;79:540-8. [PubMed]
- Wang H, Rosidi B, Perrault R, et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res 2005;65:4020-30. [PubMed]
- Simsek D, Brunet E, Wong SY, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet 2011;7:e1002080 [PubMed]
- Della-Maria J, Zhou Y, Tsai MS, et al. Human Mre11/human Rad50/Nbs1 and DNA ligase IIIalpha/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. J Biol Chem 2011;286:33845-53. [PubMed]
- Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem 2004;279:55117-26. [PubMed]
- Liang L, Deng L, Nguyen SC, et al. Human DNA ligases I and III, but not ligase IV, are required for microhomology-mediated end joining of DNA double-strand breaks. Nucleic Acids Res 2008;36:3297-310. [PubMed]
- Paul K, Wang M, Mladenov E, et al. DNA Ligases I and III Cooperate in Alternative Non-Homologous End-Joining in Vertebrates. PLoS One 2013;8:e59505 [PubMed]
- Caldecott KW. Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays 2001;23:447-55. [PubMed]
- Wang M, Wu W, Wu W, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res 2006;34:6170-82. [PubMed]
- Brown ML, Franco D, Bürkle A, et al. Role of poly(ADP-ribosyl)ation in DNA-PKcs- independent V(D)J recombination. Proc Natl Acad Sci U S A 2002;99:4532-7. [PubMed]
- Ahmed EA, de Boer P, Philippens ME, et al. Parp1-XRCC1 and the repair of DNA double strand breaks in mouse round spermatids. Mutat Res 2010;683:84-90. [PubMed]
- Mansour WY, Rhein T, Dahm-Daphi J. The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res 2010;38:6065-77. [PubMed]
- Paddock MN, Bauman AT, Higdon R, et al. Competition between PARP-1 and Ku70 control the decision between high-fidelity and mutagenic DNA repair. DNA Repair (Amst) 2011;10:338-43. [PubMed]
- Cheng Q, Barboule N, Frit P, et al. Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks. Nucleic Acids Res 2011;39:9605-19. [PubMed]
- Rosidi B, Wang M, Wu W, et al. Histone H1 functions as a stimulatory factor in backup pathways of NHEJ. Nucleic Acids Res 2008;36:1610-23. [PubMed]
- Jeggo PA, Geuting V, Löbrich M. The role of homologous recombination in radiation-induced double-strand break repair. Radiother Oncol 2011;101:7-12. [PubMed]
- Goodarzi AA, Jeggo P, Lobrich M. The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax. DNA Repair (Amst) 2010;9:1273-82. [PubMed]
- Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol 2009;16:814-8. [PubMed]
- Dinkelmann M, Spehalski E, Stoneham T, et al. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol 2009;16:808-13. [PubMed]
- Lee-Theilen M, Matthews AJ, Kelly D, et al. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat Struct Mol Biol 2011;18:75-9. [PubMed]
- Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol 2011;18:80-4. [PubMed]
- Rass E, Grabarz A, Plo I, et al. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol 2009;16:819-24. [PubMed]
- Zhuang J, Jiang G, Willers H, et al. Exonuclease function of human Mre11 promotes deletional nonhomologous end joining. J Biol Chem 2009;284:30565-73. [PubMed]
- Zhong Q, Chen CF, Chen PL, et al. BRCA1 facilitates microhomology-mediated end joining of DNA double strand breaks. J Biol Chem 2002;277:28641-7. [PubMed]
- Kuhfittig-Kulle S, Feldmann E, Odersky A, et al. The mutagenic potential of non-homologous end joining in the absence of the NHEJ core factors Ku70/80, DNA-PKcs and XRCC4-LigIV. Mutagenesis 2007;22:217-33. [PubMed]
- Elliott B, Richardson C, Jasin M. Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Mol Cell 2005;17:885-94. [PubMed]
- Bennardo N, Cheng A, Huang N, et al. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 2008;4:e1000110 [PubMed]
- Windhofer F, Wu W, Wang M, et al. Marked dependence on growth state of backup pathways of NHEJ. Int J Radiat Oncol Biol Phys 2007;68:1462-70. [PubMed]
- Zhuang J, Zhang J, Willers H, et al. Checkpoint kinase 2-mediated phosphorylation of BRCA1 regulates the fidelity of nonhomologous end-joining. Cancer Res 2006;66:1401-8. [PubMed]
- Zhang J, Willers H, Feng Z, et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol 2004;24:708-18. [PubMed]
- Zhong Q, Boyer TG, Chen PL, et al. Deficient nonhomologous end-joining activity in cell-free extracts from Brca1-null fibroblasts. Cancer Res 2002;62:3966-70. [PubMed]
- Chen L, Nievera CJ, Lee AY, et al. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem 2008;283:7713-20. [PubMed]
- Zhang Y, Gostissa M, Hildebrand DG, et al. The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Adv Immunol 2010;106:93-133. [PubMed]
- Alt FW, Zhang Y, Meng FL, et al. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 2013;152:417-29. [PubMed]
- Elliott B, Jasin M. Double-strand breaks and translocations in cancer. Cell Mol Life Sci 2002;59:373-85. [PubMed]
- Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell 2010;141:27-38. [PubMed]
- Wang JH, Gostissa M, Yan CT, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature 2009;460:231-6. [PubMed]
- Wang JH, Alt FW, Gostissa M, et al. Oncogenic transformation in the absence of Xrcc4 targets peripheral B cells that have undergone editing and switching. J Exp Med 2008;205:3079-90. [PubMed]
- Roth DB. Restraining the V(D)J recombinase. Nat Rev Immunol 2003;3:656-66. [PubMed]
- Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell 2002;109:S45-55. [PubMed]
- Jung D, Giallourakis C, Mostoslavsky R, et al. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol 2006;24:541-70. [PubMed]
- Malu S, Malshetty V, Francis D, et al. Role of non-homologous end joining in V(D)J recombination. Immunol Res 2012;54:233-46. [PubMed]
- Lieber MR, Ma Y, Pannicke U, et al. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 2004;3:817-26. [PubMed]
- Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet 2011;45:167-202. [PubMed]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol 2008;26:261-92. [PubMed]
- Xu Z, Zan H, Pone EJ, et al. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat Rev Immunol 2012;12:517-31. [PubMed]
- Nussenzweig A, Chen C, da Costa Soares V, et al. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature 1996;382:551-5. [PubMed]
- Frank KM, Sekiguchi JM, Seidl KJ, et al. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature 1998;396:173-7. [PubMed]
- Fukumura R, Araki R, Fujimori A, et al. Signal joint formation is also impaired in DNA-dependent protein kinase catalytic subunit knockout cells. J Immunol 2000;165:3883-9. [PubMed]
- Fukumura R, Araki R, Fujimori A, et al. Murine cell line SX9 bearing a mutation in the dna-pkcs gene exhibits aberrant V(D)J recombination not only in the coding joint but also in the signal joint. J Biol Chem 1998;273:13058-64. [PubMed]
- Errami A, He DM, Friedl AA, et al. XR-C1, a new CHO cell mutant which is defective in DNA-PKcs, is impaired in both V(D)J coding and signal joint formation. Nucleic Acids Res 1998;26:3146-53. [PubMed]
- Ouyang H, Nussenzweig A, Kurimasa A, et al. Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination In vivo. J Exp Med 1997;186:921-9. [PubMed]
- Moshous D, Callebaut I, de Chasseval R, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 2001;105:177-86. [PubMed]
- Buck D, Malivert L, de Chasseval R, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 2006;124:287-99. [PubMed]
- Buck D, Moshous D, de Chasseval R, et al. Severe combined immunodeficiency and microcephaly in siblings with hypomorphic mutations in DNA ligase IV. Eur J Immunol 2006;36:224-35. [PubMed]
- Enders A, Fisch P, Schwarz K, et al. A severe form of human combined immunodeficiency due to mutations in DNA ligase IV. J Immunol 2006;176:5060-8. [PubMed]
- van der Burg M, Ijspeert H, Verkaik NS, et al. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest 2009;119:91-8. [PubMed]
- Blunt T, Finnie NJ, Taccioli GE, et al. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell 1995;80:813-23. [PubMed]
- Zhu C, Bogue MA, Lim DS, et al. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell 1996;86:379-89. [PubMed]
- Taccioli GE, Amatucci AG, Beamish HJ, et al. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity 1998;9:355-66. [PubMed]
- Mombaerts P, Iacomini J, Johnson RS, et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 1992;68:869-77. [PubMed]
- Shinkai Y, Rathbun G, Lam KP, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 1992;68:855-67. [PubMed]
- Corneo B, Wendland RL, Deriano L, et al. Rag mutations reveal robust alternative end joining. Nature 2007;449:483-6. [PubMed]
- Arnal SM, Holub AJ, Salus SS, et al. Non-consensus heptamer sequences destabilize the RAG post-cleavage complex, making ends available to alternative DNA repair pathways. Nucleic Acids Res 2010;38:2944-54. [PubMed]
- Verkaik NS, Esveldt-van Lange RE, van Heemst D, et al. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur J Immunol 2002;32:701-9. [PubMed]
- Huye LE, Purugganan MM, Jiang MM, et al. Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase. Mol Cell Biol 2002;22:3460-73. [PubMed]
- Lee GS, Neiditch MB, Salus SS, et al. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell 2004;117:171-84. [PubMed]
- Deriano L, Chaumeil J, Coussens M, et al. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature 2011;471:119-23. [PubMed]
- Yan CT, Boboila C, Souza EK, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 2007;449:478-82. [PubMed]
- Han L, Yu K. Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV-deficient B cells. J Exp Med 2008;205:2745-53. [PubMed]
- Boboila C, Yan C, Wesemann DR, et al. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J Exp Med 2010;207:417-27. [PubMed]
- Kiefer K, Oshinsky J, Kim J, et al. The catalytic subunit of DNA-protein kinase (DNA-PKcs) is not required for Ig class-switch recombination. Proc Natl Acad Sci U S A 2007;104:2843-8. [PubMed]
- Bosma GC, Kim J, Urich T, et al. DNA-dependent protein kinase activity is not required for immunoglobulin class switching. J Exp Med 2002;196:1483-95. [PubMed]
- Casellas R, Nussenzweig A, Wuerffel R, et al. Ku80 is required for immunoglobulin isotype switching. EMBO J 1998;17:2404-11. [PubMed]
- Manis JP, Gu Y, Lansford R, et al. Ku70 is required for late B cell development and immunoglobulin heavy chain class switching. J Exp Med 1998;187:2081-9. [PubMed]
- Saribasak H, Maul RW, Cao Z, et al. XRCC1 suppresses somatic hypermutation and promotes alternative nonhomologous end joining in Igh genes. J Exp Med 2011;208:2209-16. [PubMed]
- Han L, Mao W, Yu K. X-ray repair cross-complementing protein 1 (XRCC1) deficiency enhances class switch recombination and is permissive for alternative end joining. Proc Natl Acad Sci U S A 2012;109:4604-8. [PubMed]
- Boboila C, Oksenych V, Gostissa M, et al. Robust chromosomal DNA repair via alternative end-joining in the absence of X-ray repair cross-complementing protein 1 (XRCC1). Proc Natl Acad Sci U S A 2012;109:2473-8. [PubMed]
- Simsek D, Furda A, Gao Y, et al. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature 2011;471:245-8. [PubMed]
- Robert I, Dantzer F, Reina-San-Martin B. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J Exp Med 2009;206:1047-56. [PubMed]
- Shockett P, Stavnezer J. Inhibitors of poly (ADP-ribose) polymerase increase antibody class switching. J Immunol 1993;151:6962-76. [PubMed]
- Audebert M, Salles B, Calsou P. Effect of double-strand break DNA sequence on the PARP-1 NHEJ pathway. Biochem Biophys Res Commun 2008;369:982-8. [PubMed]
- Sandoval A, Labhart P. High G/C content of cohesive overhangs renders DNA end joining Ku-independent. DNA Repair (Amst) 2004;3:13-21. [PubMed]
- Zhu C, Mills KD, Ferguson DO, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell 2002;109:811-21. [PubMed]
- Du L, Peng R, Björkman A, et al. Cernunnos influences human immunoglobulin class switch recombination and may be associated with B cell lymphomagenesis. J Exp Med 2012;209:291-305. [PubMed]
- Sattler M, Verma S, Shrikhande G, et al. The BCR/ABL tyrosine kinase induces production of reactive oxygen species in hematopoietic cells. J Biol Chem 2000;275:24273-8. [PubMed]
- Sallmyr A, Fan J, Rassool FV. Genomic instability in myeloid malignancies: increased reactive oxygen species (ROS), DNA double strand breaks (DSBs) and error-prone repair. Cancer Lett 2008;270:1-9. [PubMed]
- Poplawski T, Blasiak J. BCR/ABL downregulates DNA-PK(CS)-dependent and upregulates backup non-homologous end joining in leukemic cells. Mol Biol Rep 2010;37:2309-15. [PubMed]
- Mattarucchi E, Guerini V, Rambaldi A, et al. Microhomologies and interspersed repeat elements at genomic breakpoints in chronic myeloid leukemia. Genes Chromosomes Cancer 2008;47:625-32. [PubMed]
- Sallmyr A, Tomkinson AE, Rassool FV. Up-regulation of WRN and DNA ligase IIIalpha in chronic myeloid leukemia: consequences for the repair of DNA double-strand breaks. Blood 2008;112:1413-23. [PubMed]
- Fan J, Li L, Small D, et al. Cells expressing FLT3/ITD mutations exhibit elevated repair errors generated through alternative NHEJ pathways: implications for genomic instability and therapy. Blood 2010;116:5298-305. [PubMed]
- Meek K, Gupta S, Ramsden DA, et al. The DNA-dependent protein kinase: the director at the end. Immunol Rev 2004;200:132-41. [PubMed]
- Weterings E, Chen DJ. DNA-dependent protein kinase in nonhomologous end joining: a lock with multiple keys? J Cell Biol 2007;179:183-6. [PubMed]
- Soutoglou E, Dorn JF, Sengupta K, et al. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol 2007;9:675-82. [PubMed]
- Difilippantonio MJ, Zhu J, Chen HT, et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 2000;404:510-4. [PubMed]
- Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol 2007;9:978-81. [PubMed]
- Bentley J, Diggle CP, Harnden P, et al. DNA double strand break repair in human bladder cancer is error prone and involves microhomology-associated end-joining. Nucleic Acids Res 2004;32:5249-59. [PubMed]
- D’Adda di Fagagna F, Reaper PM, Clay-Farrace L, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003;426:194-8. [PubMed]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol 2003;13:1549-56. [PubMed]
- de Lange T. How telomeres solve the end-protection problem. Science 2009;326:948-52. [PubMed]
- Espejel S, Franco S, Rodríguez-Perales S, et al. Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J 2002;21:2207-19. [PubMed]
- Fisher TS, Zakian VA. Ku: a multifunctional protein involved in telomere maintenance. DNA Repair (Amst) 2005;4:1215-26. [PubMed]
- Gilley D, Tanaka H, Hande MP, et al. DNA-PKcs is critical for telomere capping. Proc Natl Acad Sci U S A 2001;98:15084-8. [PubMed]
- Goytisolo FA, Samper E, Edmonson S, et al. The absence of the dna-dependent protein kinase catalytic subunit in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol Cell Biol 2001;21:3642-51. [PubMed]
- Hsu HL, Gilley D, Galande SA, et al. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev 2000;14:2807-12. [PubMed]
- Bombarde O, Boby C, Gomez D, et al. TRF2/RAP1 and DNA-PK mediate a double protection against joining at telomeric ends. EMBO J 2010;29:1573-84. [PubMed]
- Hsu HL, Gilley D, Blackburn EH, et al. Ku is associated with the telomere in mammals. Proc Natl Acad Sci U S A 1999;96:12454-8. [PubMed]
- Maser RS, Wong KK, Sahin E, et al. DNA-dependent protein kinase catalytic subunit is not required for dysfunctional telomere fusion and checkpoint response in the telomerase-deficient mouse. Mol Cell Biol 2007;27:2253-65. [PubMed]
- Rai R, Zheng H, He H, et al. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J 2010;29:2598-610. [PubMed]
- Oh S, Wang Y, Zimbric J, et al. Human LIGIV is synthetically lethal with the loss of Rad54B-dependent recombination and is required for certain chromosome fusion events induced by telomere dysfunction. Nucleic Acids Res 2013;41:1734-49. [PubMed]
- Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917-21. [PubMed]
- Chen X, Zhong S, Zhu X, et al. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer Res 2008;68:3169-77. [PubMed]
- Tobin LA, Robert C, Rapoport AP, et al. Targeting abnormal DNA double-strand break repair in tyrosine kinase inhibitor-resistant chronic myeloid leukemias. Oncogene 2013;32:1784-93. [PubMed]
- Tobin LA, Robert C, Nagaria P, et al. Targeting abnormal DNA repair in therapy-resistant breast cancers. Mol Cancer Res 2012;10:96-107. [PubMed]
- Fattah F, Lee EH, Weisensel N, et al. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet 2010;6:e1000855 [PubMed]
- Bothmer A, Robbiani DF, Feldhahn N, et al. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med 2010;207:855-65. [PubMed]
- Bothmer A, Robbiani DF, Di Virgilio M, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell 2011;42:319-29. [PubMed]
- Helmink BA, Tubbs AT, Dorsett Y, et al. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature 2011;469:245-9. [PubMed]
- Lundberg R, Mavinakere M, Campbell C. Deficient DNA end joining activity in extracts from fanconi anemia fibroblasts. J Biol Chem 2001;276:9543-9. [PubMed]
- Perrault R, Wang H, Wang M, et al. Backup pathways of NHEJ are suppressed by DNA-PK. J Cell Biochem 2004;92:781-94. [PubMed]
- Callén E, Jankovic M, Difilippantonio S, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell 2007;130:63-75. [PubMed]
- Bredemeyer AL, Sharma GG, Huang CY, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature 2006;442:466-70. [PubMed]



