
Inhibit a kinase to degrade a histone demethylase: a candidate therapeutic approach in glioblastoma
Lysine-specific histone demethylase 1A (KDM1A; more commonly known as LSD1), the first enzyme discovered to possess histone demethylase activity, removes monomethyl and dimethyl groups from histone H3 lysine 4 (H3K4) (1). It is generally considered a transcription repressor being a key component of corepressor complexes such as CoREST and NuRD (2-4). High level expression of KDM1A protein is observed in multiple cancer types, including poor prognosis sub-groups of prostate, lung, brain and breast cancer, as well as in certain hematological malignancies. Pharmacologic inhibitors of KDM1A are currently in early phase clinical trials (4). While transcriptional regulation of KDM1A undoubtedly plays a significant role in specifying cellular levels of KDM1A protein, it has recently become clear that post-translational mechanisms are also important. For example, the differentiation of embryonic stem cells towards neuronal cell fate is regulated by the E3-ubiquitin ligase PHF15 (also known as JADE2), by facilitating though polyubiquitination proteasomal degradation of KDM1A (5). On the contrary, the deubiquitinase USP28 stabilizes KDM1A protein, and its expression correlates with that of KDM1A in cancer cell lines and patient breast cancer samples (6). Whether PHF15 and USP28 are the only post-translational regulators of KDM1A levels has not been clear. Likewise, whether the cellular pathway of KDM1A degradation can be targeted for potential therapeutic benefit is also an open issue. Both these questions have been addressed in a recent, elegant and comprehensive study from Aidong Zhou, Kangyu Lin, Suyun Huang and colleagues in the setting of glioblastoma (7). Glioblastoma is the most common type of malignant brain tumor, with very poor survival. The frequent therapeutic resistance and recurrence following resection indicate a need for new treatment approaches (8).
Given previous reports of functional roles for KDM1A and glycogen synthase kinase 3 (GSK3) in glioblastoma, the authors set out to investigate whether the two enzymes might cooperate in the same cellular pathway. Phosphorylation by GSK3 has previously been reported to promote ubiquitination and degradation of target proteins. GSK3 was initially described as one of several protein kinases that phosphorylate glycogen synthase, involved in the last step in glycogen synthesis. It exists as two isoforms (GSK3A and GSK3B) encoded by separate genes and has many important cellular roles. In contrast to most kinases, GSK3 is active under resting conditions and its downstream targets are typically held in an inactive state through GSK3-mediated phosphorylation. GSK3 kinase activity itself is inhibited by diverse activating stimuli, including insulin signaling via the phosphoinositide-3-kinase/Akt pathway. Dysregulation of GSK3 has been implicated in various human diseases such as diabetes, cancer and neurodegenerative diseases (9). GSK3B is a key component of the Wnt signaling pathway, forming a complex with adenomatous polyposis coli (APC) and axin to phosphorylate β-catenin, resulting in its degradation. GSK3 is also involved in Alzheimer’s disease through its association with presenilin and its phosphorylation of the microtubule-associated protein tau (9). More recent evidence has revealed significant effects on epigenetic regulators, with several examples of GSK3 modifying histone deacetylases to regulate their stability and activity (10,11).
Zhou, Lin and colleagues first examined multiple glioblastoma cell lines and found a robust correlation of nuclear KDM1A protein levels with those of GSK3B. Making use of genetic knockdown, knockout and forced expression experimental approaches, they discovered that cellular levels of KDM1A protein, but not transcripts, are directly regulated by GSK3B. Indeed, blockade of proteasomal degradation was sufficient to restore KDM1A protein levels following GSK3B knockdown, raising a question as to whether GSK3B directly regulates KDM1A protein stability. This was confirmed in preliminary experiments where GSK3B was observed to stabilize KDM1A by decreasing its ubiquitination.
Next, candidate GSK3B phosphorylation sites [i.e., those harboring the canonical (S/T)XXX(S/T) motif] were identified in KDM1A in silico, and through a targeted mutation approach, Ser683 was identified as the residue that, when phosphorylated by GSK3B, reduced KDM1A ubiquitination. GSK3 substrates first require a priming phosphorylation at a site located four residues carboxy-terminal to the GSK3 phosphorylation site: thus, the most common target for GSK3B phosphorylation is the phosphorylated sequence (S/T)XXX(S/T) (P) (9). Using an in vitro kinase assay the authors showed the relevant priming kinase to be casein kinase 1 alpha (CSNK1A1). Pharmacological inhibition of either CSNK1A1 (with D4476) or GSK3 (with lithium chloride) decreased endogenous pKDM1A-Ser683 levels.
In view of the observation that GSK3B-mediated phosphorylation of KDM1A led to a decrease in KDM1A ubiquitination, the authors next examined a panel of deubiquitinases looking for one that could function in cooperation with GSK3B. Of those tested, only USP22 was found directly to deubiquitinate KDM1A. Forced expression of USP22 substantially decreased KDM1A ubiquitination and increased its stability, whereas USP22 knockdown decreased KDM1A protein levels without affecting KDM1A mRNA. Moreover, in primary patient glioblastoma samples KDM1A and USP22 co-localized frequently in the nuclei, and were shown to physically interact in cell lines though the amine oxidase domain of KDM1A (the same site of interaction of KDM1A with GSK3B). Knockdown of GSK3B or expression of a S683A KDM1A mutant significantly decreased the interaction between USP22 and KDM1A, suggesting that Ser683 phosphorylated KDM1A is able to associate more efficiently with the USP22 deubiquitinase than un-phosphorylated KDM1A, providing a mechanistic explanation for the experimental observations (Figure 1).
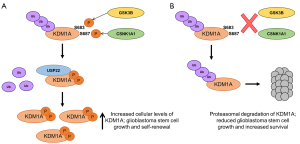
That KDM1A protein levels are regulated to functional effect in glioblastoma cells by GSK3B and USP22 was confirmed using a genetic knockdown approach. Knockdown of GSK3B and USP22 gave similar results to KDM1A knockdown with regard to accumulation of dimethyl- and trimethyl-H3K4 histone methylation marks at the promoters of KDM1A target genes, and increased transcript expression. Furthermore, the frequency of neurosphere forming cells in a glioblastoma stem cell (GSC) line was reduced by knockdown of either KDM1A or USP22, as was expression of stem cell markers such as Nestin, OCT4 and CD133; in both instances the loss of neurosphere-forming potential was rescued by re-expression of KDM1A.
The investigators next made use of mouse models to investigate whether GSK3B and USP22 regulate the tumorigenicity of GSCs through KDM1A in vivo. Depletion of KDM1A, USP22 or GSK3B transcripts in a GSC line abrogated brain tumor formation and once more the inhibitory effects could be rescued by forced expression of KDM1A. The inhibitory effect of GSK3B transcript depletion was rescued by expression of the phosphomimetic KDM1A-S683D mutant but not the non-phosphorylatable KDM1A-S683A mutant, suggesting that GSK3B-dependent phosphorylation of KDM1A is required for the tumorigenicity of GSCs in vivo. The observation of an immunohistochemical correlation between nuclear KDM1A, GSK3B and USP22 in 96 human high grade glioblastomas (WHO grade IV) sections is consistent with this being an important mechanism in patients, and led on to the final, pre-clinical stage of the study.
Several GSK3 inhibitors are in development with a focus on Alzheimer’s disease. Among them, tideglusib (a non-ATP-competitive inhibitor) decreases tau hyper-phosphorylation, lowers brain amyloid plaque levels, improves learning and memory, and prevents neuronal loss in animal models. Early phase trials confirmed safety and larger clinical trials are underway (12). In glioblastoma treatment, temozolomide (a DNA alkylating agent) is the current standard of care but patients typically exhibit high rates of recurrence (8). Zhou, Lin and colleagues tested whether tideglusib could reduce glioblastoma cell viability and sensitize them to temozolomide. In keeping with their hypothesis, tideglusib induced KDM1A degradation, reduced neurosphere formation and decreased expression of the neural stem cell markers, without apparent toxicity to normal human astrocytes. In vivo, tideglusib alone significantly decreased tumor size, but when combined with temozolomide tumor formation was almost entirely abrogated. Importantly, in glioblastoma-bearing mice tideglusib alone had a better effect on survival than temozolomide alone, an effect that that was potentiated with the combination of the two drugs. Although all mice finally succumbed to disease, these data provide a powerful rationale for investigating this therapeutic combination in clinical trials, especially given the apparent good penetration of the blood-brain barrier by tideglusib.
Together these data demonstrate that KDM1A contributes to the maintenance of stemness in glioblastoma cells and that its cellular levels can be targeted for degradation through inhibition of the kinase GSK3. The resulting loss of phosphorylation at Ser683 reduces physical association of the USP22 deubiquitinase, enhancing proteasomal destruction. This study opens up a new approach to therapeutic targeting of KDM1A, over and above pharmacologic inhibition of its enzymatic activity. Key questions remain including whether the same GSK3B-USP22-KDM1A regulatory axis operates in other tumor types where KDM1A overexpression is observed. For instance, GSK3 activity has been shown to be essential for maintenance of a subset of leukemias driven by mixed-lineage leukemia (MLL) translocation (13), the same leukemia subset that is dependent on expression of KDM1A and responds to KDM1A inhibition (14). Also, in breast cancer another deubiquitinase USP28 has already been linked to KDM1A accumulation (6), but the mechanism upstream remains unclear. Whether GSK3B plays a role in these and other KDM1A-dependent tumors is an interesting question for future study, especially given KDM1A has been shown to be a good therapeutic target, and with different pharmacological inhibitors currently in the first steps of clinical trials, in particular in the settings of acute myeloid leukaemia and small-cell lung cancer (4).
Acknowledgments
Funding: A Maiques-Diaz and TC Somervaille are supported by Cancer Research UK Grant Number C5759/A12328.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Chen Qian (Center for Inflammation & Epigenetics, Houston Methodist Hospital Research Institute, Houston, USA).
Conflicts of Interest: TC Somervaille has research collaborations with Oryzon Genomics and consults for Imago Biosciences. The other author has no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004;119:941-53. [Crossref] [PubMed]
- Shi YJ, Matson C, Lan F, et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 2005;19:857-64. [Crossref] [PubMed]
- Wang Y, Zhang H, Chen Y, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 2009;138:660-72. [Crossref] [PubMed]
- Maiques-Diaz A, Somervaille TC. LSD1: biologic roles and therapeutic targeting. Epigenomics 2016;8:1103-16. [Crossref] [PubMed]
- Han X, Gui B, Xiong C, et al. Destabilizing LSD1 by Jade-2 promotes neurogenesis: an antibraking system in neural development. Mol Cell 2014;55:482-94. [Crossref] [PubMed]
- Wu Y, Wang Y, Yang XH, et al. The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell Rep 2013;5:224-36. [Crossref] [PubMed]
- Zhou A, Lin K, Zhang S, et al. Nuclear GSK3β promotes tumorigenesis by phosphorylating KDM1A and inducing its deubiquitylation by USP22. Nat Cell Biol 2016;18:954-66. [Crossref] [PubMed]
- Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell 2012;149:36-47. [Crossref] [PubMed]
- Cohen P, Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol 2001;2:769-76. [Crossref] [PubMed]
- Bardai FH, D'Mello SR. Selective toxicity by HDAC3 in neurons: regulation by Akt and GSK3beta. J Neurosci 2011;31:1746-51. [Crossref] [PubMed]
- Cernotta N, Clocchiatti A, Florean C, et al. Ubiquitin-dependent degradation of HDAC4, a new regulator of random cell motility. Mol Biol Cell 2011;22:278-89. [Crossref] [PubMed]
- Liu H, Wang L, Su W, et al. Advances in recent patent and clinical trial drug development for Alzheimer's disease. Pharm Pat Anal 2014;3:429-47. [Crossref] [PubMed]
- Wang Z, Smith KS, Murphy M, et al. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature 2008;455:1205-9. [Crossref] [PubMed]
- Harris WJ, Huang X, Lynch JT, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 2012;21:473-87. [Crossref] [PubMed]


