
A microwell-based near-infrared fluorescence assay of DNA methylation
Introduction
DNA methylation is one of the most common epigenetic modifications, and has profound effects on the mammalian genome (1). DNA methylation has an important regulatory role in many biological processes, such as embryonic development, gene transcription, chromosome structure modulation, X chromosome inactivation, and genomic imprinting. Abnormal methylation patterns are often associated with the incidence of diseases, such as immune deficiency, mental disorders, cancer, and renal oxidative stress (2,3). Hypermethylation of CpG islands in promoter regions have frequently been detected in tumor cells (4,5), and have a close relationship with transcriptional repression and inactivation of tumor suppressor genes in cancer. Therefore, DNA methylation provides a potential target for cancer diagnosis and therapy (6). The study of DNA methylation, especially in combination with gene transcription regulation, should provide a deeper understanding of the epigenetic regulatory mechanism of DNA methylation in many biological processes.
To study the regulatory effects of DNA methylation on gene transcription, DNA methylation in the genome must be accurately detected. At present, genomic DNA (gDNA) methylation can be detected with various methods (7,8). Among them, the most common are those based on bisulfite treatment, such as bisulfite-treated DNA sequencing (9), quantum dot (QD)-based method (10), methylation-specific polymerase chain reaction (MSP) (11), and DNA microarray hybridization (12). However, these methods are susceptible to the efficiency of bisulfite treatment, because incomplete and inconsistent conversion of cytosine to uracil may produce false methylations (9). In addition, the bisulfite treatment process is detrimental to DNA and is time-consuming (~16 h) (13).
Recently, some bisulfite-independent methods have been developed (14-18), for example, the methylated DNA immunoprecipitation (MeDIP)-based methods (19,20). DNA enriched by MeDIP can be analyzed by locus-specific PCR, by global profiling techniques, such as DNA microarrays (21,22), and by next-generation DNA sequencing (MeDIP-seq) (23,24). However, MeDIP-based methods often present several disadvantages, such as the time-consuming immunoprecipitation processes, high cost of high-throughput DNA sequencing, and expensive equipment. There also reported some new methods related to nanoparticles or nanoclusters, such as the method based on the FRET mechanism between upconversion nanoparticles and gold nanorods (25) and a colorimetric and fluorimetric technique for direct detection of DNA methylation based on silver nanoclusters (26). Kermani et al. developed a methyltransferase activity assay by employing DNA-templated silver nanoclusters without using restriction enzymes, which showed a convenient reproducibility and sensitivity indicating its potential for the determination of methyltransferase activity (27). However, the nanoparticles or nanoclusters used in these methods provide a new view for the methylation detection.
Near-infrared fluorescence (NIRF)-base method have developed rapidly in recent years. The NIRF has the exciting wave-length from 700 to 900 nm. In comparison with traditional visible fluorescence, NIRF has several significant advantages, including high sensitivity, high signal-to-noise (S/N) ratio and deep tissue penetration capability (28-30). Therefore, NIRF techniques have been rapidly applied to in vitro assays of various biomolecules. For example, our lab recently applied NIRF techniques to the detections of gene transcription (31), protein expression (32) and transcription factor activity (33,34) and developed several new approaches for assaying normal biomolecules (31,33,34). We found that the common solid-phase media that are used for biochemical detections, including glass slides (33,34), microwell platesJeny (31) and PVDF (32) and nylon membranes (35) had very low autofluorescence at the exciting wavelength of some NIRF dyes such as IRDye® 800CW (exciting/emission wavelength: 774/789 nm). Therefore, detections based on these NIRF dyes obtained high sensitivity due to the minimal background interference.
In this study, we applied the NIRF techniques to the detection of DNA methylation and developed a NIRF-based method for assaying DNA methylation, a well-based near-infrared fluorescence assay (W-NIFA). The procedure of this method is straightforward that detects DNA methylation by two steps, including hybridizing the sheared gDNA to a probe-coupled microwell plate and detecting the methylated cytosines in captured DNAs by a immunological process. This study validated this method by detecting methylation of nine loci in promoters of three genes in three cancer cell lines with this method and bisulfite sequencing PCR. This method has several advantages such as circumvents some cumbersome and time-consuming procedures of other traditional methods, such as bisulfite treatment, primer design for MSP, and immunoprecipitation of methylated DNA. In addition, this method was designed to detect methylation of both DNA strands and is suitable for the detection of the methylation levels of interested genomic locations.
Methods
Preparation of the DNA-coupled plate, gDNA, total RNA
The oligonucleotides used in this paper were synthesized by Sangon Biotechnology (Shanghai, China), and the sequence is listed in Table 1. The probe-coupled 96-well microplate was prepared as previously described (36) with slightly modified as follows: the DNA-BIND white 96-well polystyrene microplate (Corning Inc. Life Sciences, Tewksbury, MA, USA) was used as solid-phase support for immobilizing oligonucleotide probe in this study. The surface of DNA-BIND microplate has covalently linked N-oxysuccinimide (NOS) groups that react with nucleophiles such as primary amines at slightly alkaline pH. The amino-modified oligonucleotide probes (Table 1) were dissolved in oligo binding buffer (50 mM Na3PO4, pH 8.5, 1 mM EDTA) at the concentration of 100 µM. Before coupling to a microplate, oligonucleotides were diluted with OBB at a final concentration of 0.25 µM, then added to the microplate at 100 μL per well. The microplate was incubated overnight at 4 °C in advance before used. Then washed 3 times with OBB to remove uncoupled DNA and blocked with blocking solution (3% BSA in OBB) at 37 °C for 30 min in volume of 200 μL per well. Cells, including HepG2, LOVO, K562 and HFL-1, were purchased from China Center for Type Culture Collection (Shanghai, China), and cultured according to the manufacturer’s instructions. gDNA was isolated with AxyPrep™ Blood Genomic DNA Miniprep Kit (Axygen Scientific), according to the manufacturer’s protocols. Total RNA from cells was extracted using TRIzol reagent (TaKaRa) as per the manufacturer’s instructions. The quality and quantity of the extracted gDNA and total RNA were evaluated by agarose gel electrophoresis and NanoDrop® ND-1000 Spectrophotometer (Thermo Fisher), respectively. The gDNA detected with the probe-coupled microplate was sheared into small fragments around 100 bp by sonication (sonication condition: working time 28 s, break time 8 s, the total working time around 20 min at 4 °C).
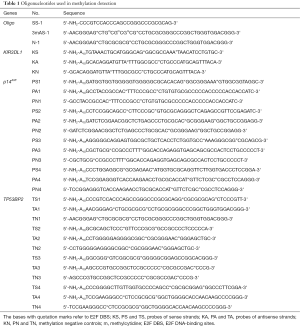
Full table
Hybridization of gDNA to the DNA-coupled plate
The sonicated gDNA was heated at 95 °C for 5 min, and immediately chilled on ice. The synthesized complementary oligonucleotide probes (mAS-1, N-1 in Table 1, Sangon Biotech) or the denatured gDNA was added to a temperature of melting temperature (Tm) minus 25 °C (Tm-25 °C)-heated hybridization solution {5 × SSC (1 × SSC: 0.15 M NaCl, 0.015 M sodium citrate, pH 7.0), 0.02% SDS, 0.1% N-lauroylsarcosine, 1% blocking reagent [phosphate-buffered saline (PBS), pH 7.4, 1% BSA], freshly prepared}, and 100 µL (75–200 µL of recommended working volume) per well (~300 ng gDNA per well) was added into wells of the plate and incubated for 5–6 h at a temperature of Tm-25 °C. To prepare a methylation-negative control DNA for each locus, a synthesized complementary oligonucleotide (N-1, KN, PN and TN in Table 1) was hybridized to the immobilized probes (Table 1) during this step. After hybridization, the plate was washed once with 2 × SSC, 0.1% SDS at room temperature (RT), once with 0.5 × SSC, 0.1% SDS preheated to Tm-25 °C. The plate was briefly washed once with with PBST (PBS, pH 7.4, 0.05% Tween 20).
Detection of DNA methylation
DNA methylation [including the methylation of E2F DNA-binding sites (E2F DBS)] was detected on the oligonucleotide probes-hybridized plate or the gDNA-hybridized plate using the following two steps: (I) mouse anti-5-methylcytosine (Epigentek, NY), diluted 1:2,000 in antibody dilution buffer (PBS, pH 7.4, 0.1% BSA), was added to the plate and incubated at RT for 2–3 h; (II) IRDye® 800CW-conjugated goat (polyclonal) anti-mouse IgG (Li-Cor, NE), diluted 1:2,000 in antibody dilution buffer, was added to plate and incubated at RT for 1 h. All solutions were added to plate at 100 µL per well, and after each step, the plate was washed 3 times with PBST. Finally, the plate was scanned with Odyssey Infrared Imaging System (Li-Cor, NE) at the channel of 800 nm. The intensity of images was quantified using the Odyssey Infrared Imaging System.
Detection of DNA methylation with bisulfite sequencing PCR
The gDNA samples were subjected to bisulfite modification by using the EpiTect Bisulfite Kit (Qiagen, #59104) according to the manufacturer’s recommendations. After treatment of bisulfite, cytosine residues of gDNA were converts into uracil, and the 5-methylcytosine residues were unaffected. Thus, bisulfite treatment introduces specific changes in the DNA sequence that depend on the methylation status of individual cytosine residues. Then the promoter regions of KIR3DL1, p14ARF and TP53BP2 (ASPP2) were amplified from the bisulfite-treated gDNA with the primers listed in Table 2. The results were detected by the agarose gel electrophoresis. Bisulfite sequencing were performed with at least eight individual clones by using ABI3730 automatic DNA sequencer (Applied Biosystems). The promoter-associated CpG islands were searched online with CpG Island Searcher (37) using the NCBI original sequence. The results of bisulfite sequencing PCR were analyzed online with Bisulfite Sequencing DNA Methylation Analysis software (BISMA) with FASTA format (38).

Full table
Detection of gene expression with quantitative PCR
Complementary DNA (cDNA) was synthesized from 1 μg of total RNA as a template using a reverse transcriptase kit (TaKaRa), following manufacturer’s instructions. Quantitative PCR was performed with Fast SYBR Master Mix (Applied Biosystems, UK) using primers listed in Table 2 on a StepOne™ Plus instrument (Applied Biosystems). The PCR reaction consisted of 10 µL 2× Fast SYBR Master Mix, 1 µL cDNA, 0.4 µL 10 µM forward primer (Table 2), 0.4 µL 10 µM reverse primer (Table 2), and 8.2 µL water. The PCR reaction was: 95 °C 10 min; 40 cycles of 95 °C 15 s, 58 °C (TP53BP2 and p14ARF) or 60 °C (KIR3DL1) 1 min; and 72 °C 5 min. A melting curve stage was performed after the completion of PCR. The data for the target genes were normalized to that of a housekeeping gene (gapdh).
Results
Analytical principle of this method
The analytical principle of this method for detection of DNA methylation is schematically illustrated in Figure 1. With the prefabricated probe-coupled microwell plate, the detection process of this method mainly consists of two steps: (I) hybridize the sonicated gDNA with probe-coupled microwell plate; (II) detect DNA methylation with a immunological procedure that used anti-methylated cytosine antibody and a IRDye® 800CW-labeled second antibody. To evaluate this method, the methylations of nine loci in promoters of three genes, KIR3DL1, p14ARF and TP53BP2, in three cancer cell lines were detected with this method.
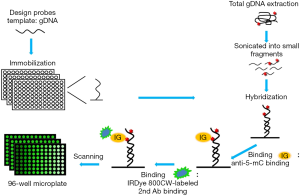
Quantitative detection of methylation
To investigate the specificity and sensitivity of this method, a standard curve was prepared using oligonucleotides contained three methylated cytosines (oligo 3mAS-1 in Table 1) in a series of concentration (0–30 nM of 0–9 pmoL methylated cytosines in Figure 2) and oligonucleotide with no methylcytidine modified as the methylation negative control (oligo N-1, columns 12). The oligonucleotides were hybridized to a complementary probes (oligo SS-1 at the concentration of 0.25 µM coupled to a plate) and detecting with the immunological procedure. The results revealed that the standard curve with R2 >0.99 could be plotted between 0 to 7.2 pmoL of the methylated cytosines and as low as 0.9 pmoL (3 nM × 100 µL ×3) of methylated cytosines could be detected. The methylation negative control in quadruplets produced no NIRF signal showed a good specificity of this method.

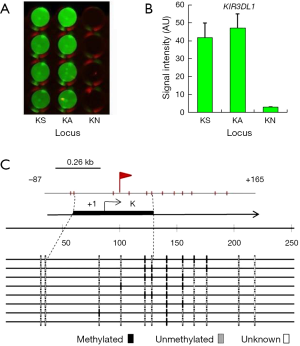
Detection of KIR3DL1 promoter methylation
The results of the detection of methylation of the sole site containing the E2F DBS in the KIR3DL1 promoter in K562 cell are shown in Figure 3. This site was detected with the methylated signal in K562 cells and no signal had been detected with the negative control dsDNA probe (N) in W-NIFA (Figure 3A,B). The bisulfite sequencing PCR result (Figure 3C) revealed that 22.5% CpG [mCG/(CG × clone number)%=9/(5×8)%=22.5%] in ~100 bp of the E2F DBS was methylated in K562 cells.
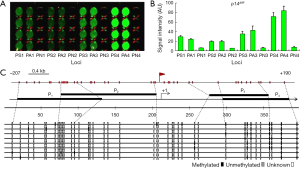
Detection of p14ARF promoter methylation
The results of the detection of the methylation of the four loci containing the E2F DBSs in the p14ARF promoter in LOVO cells are shown in Figure 4. The four loci in the p14ARF promoter were all methylated in LOVO cells, but the degree of methylation of these sites were not identical. It was clear that the 4th locus had the highest methylation level and that the 2nd locus had the lowest methylation level (Figure 4A,B). The bisulfite sequencing PCR also revealed the similar variance of methylation levels at four loci (Figure 4C). In these loci, the 4th locus had the highest methylation level, 52.5% methylated CpG [mCG/(CG × clone number)%=42/(10×8)%=52.5%], the 3rd locus had the medium methylation level, 31.9% methylated CpG [mCG/(CG × clone number)%=23/(9×8)%=31.9%], and the 1st and 2nd loci had the lowest methylation level, 6.25% and 3.13% methylated CpG, respectively.
Detection of TP53BP2 promoter methylation
The method was also used to detect the methylation of four loci of TP53BP2 promoter in HepG2 cells. The results are displayed in Figure 5, which demonstrated that these loci were methylated in various degrees in HepG2 cells (Figure 5A,B). The similar methylation levels were also detected by bisulfite sequencing PCR (Figure 5C). In these loci, the 3rd loci showed the highest NIRF intensity and methylated CpGs. 6.62% [mCG/(CG × clone number)%=9/(17×8)%=6.62%] CpGs in ~100 bp around capture probe were methylated. The methylation degrees of the 1st and 2nd loci were similar; the bisulfite sequencing PCR revealed that 2.21% and 2.98% CpGs of two loci were methylated in HepG2 cells. Both methods detected the lowest methylation of the 4th locus, the lowest signal detected by W-NIFA and 0.74% methylated CpGs detected by bisulfite sequencing.
Correlation analysis of methylation levels
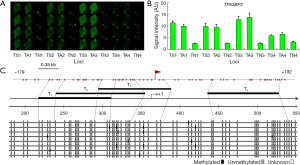
To evaluate the accuracy of W-NIFA detection, correlation analysis of methylation levels of sense and antisense strands were performed between W-NIFA and bisulfite sequencing PCR results. It was revealed that the methylations of the antisense-strands of three genes, KIR3DL1, p14ARF and TP53BP2, were strongly correlated (R2 =0.886) (Figure 6A,B). The similar correlation were obtained to the sense strands of three genes (R2 =0.944) (Figure 6C,D).
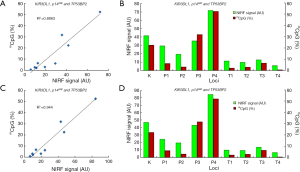
Detection of gene expression
To investigate the effect of DNA methylation on gene expression, the transcription of the four genes in their reported positive and negative cells was detected using quantitative PCR. The results are shown in Figure 7. The transcriptions of KIR3DL1 in K562 cells, p14ARF in LOVO cells, TP53BP2 in HepG2 cells were significantly lower than their transcriptions in HFL-1 cells. Human lung fibroblasts HFL-1 was used as a methylation-negative control cell in this study due to the non-methylation of the promoters of three genes in this cell lines (39,40).
Discussion
This study describes a method for detecting DNA methylation. This method was evaluated by detecting the methylation of synthesized oligonucleotides and genomic DNAs from cancer cell lines. The methylation detection results were confirmed by bisulfite sequencing PCR. The methylation detection results were also in agreement with the previous studies. For example, the CpGs in the 3rd locus of p14ARF promoter were found to hypermethylated in LOVO cells (41,42). The methylation of KIR3DL1 promoter in K562 cells and its demethylation-reversible transcription repression of the KIR3DL1 gene was reported by a previous study (43).
At present, gDNA methylation can be detected with various methods, the most common methods are MS-PCR, bisulfite sequencing and MS-HRM. The MS-PCR method has the advantages of being highly sensitive (able to detect one methylated allele in a population of more than 1,000 unmethylated alleles), and can be used on DNA samples of limited quantity and quality. However, MSP is not quantitative. In addition, the design of primers is essential for this method (7); Bisulfite sequencing is the gold standard for the methylation detection. However, this method is susceptible to the efficiency of bisulfite treatment, because incomplete and inconsistent conversion of cytosine to uracil may produce false methylations. In addition, the bisulfite treatment process is detrimental to DNA and is time-consuming (~16 h). MS-HRM protocol enables clear identification of a methylated PCR product in an unmethylated background at both 1% and 0.1% dilution points, with sensitivity similar to that of methylation-specific PCR (MSP). Furthermore, MS-HRM-based methylation screening is cost, labor and time efficient in contrast to direct bisulfite sequencing. However, MS-HRM protocol does not reveal the methylation status of the CpG sites within the primer binding sites. This disadvantage can only be overcome by redesigning the primer set (44).
Compared with the above methods, the method in this study has several advantages. First, this method circumvents some cumbersome and time-consuming procedures of other traditional methods, such as bisulfite treatment, primer design for MSP, and immunoprecipitation of methylated DNA. For example, the hypermethylation of the p14ARF promoter was detected in many colorectal carcinoma cell lines (SW48, RKO, DLD-1 and LOVO) (41,42,45,46). However, all these studies detected p14ARF methylation using MSP with a pair of primers designed by Esteller et al., which only detected the 3rd E2F DBS (47). Therefore, the methylations of the other three loci of this promoter were not detected. However, this method detected the hypermethylation of four loci of this promoter.
Second, this method was designed to detect methylation of both DNA strands. The exact methylation symmetry between the complementary strands of individual DNA molecules have been extensively reported in previous studies, and also confirmed by the results of methylation detection of nine loci in the promoters of three genes in three tumor cell lines in this study. However, the presence of asymmetric methylation in genome was also found (48,49). The asymmetric methylation detected in the human IGF2-H19 imprinted region was associated with tissue-specific disruption of H19 genomic imprinting in the fetal brain (49). The relative importance of symmetric and asymmetric methylation in the regulation of gene expression is still unknown. Therefore, this method was designed to detect methylation of both DNA strands. Last, the combination of NIRF make this method with high sensitivity, high S/N ratio and deep tissue penetration capability, and as low as 0.9 pmoL (3 nM × 100 µL ×3) of methylated cytosines could be detected.
However, the method described here still has its limitations. For example, this method can not be used to determine the definite methylation pattern of a genomic location. In addition, antibody and the specialized detection system were required in this study which increase the detection cost.
It should be pointed out that each locus detected in this study contained DNA-binding sites of transcription factor E2F (Table 1) and the genes KIR3DL1, p14ARF and TP53BP2 (ASPP2) are the known target genes of E2F (43,50,51). Many previous studies demonstrated that the methylation of the promoters of these genes inversely correlated with the expression levels of these genes (43,50-53). This study detected the methylations of nine loci in the promoters of these genes in three cancer cell lines with this method, the results were supported by bisulfite sequencing PCR and previous studies. The transcription detection revealed that the transcriptions of these genes in the detected cell lines were significantly reduced, which is in agreement with the previous studies that reported that the methylation of the promoters of these genes repressed their expressions in LOVO, HepG2 and K562 cells (39,41,43,52).
This study realized a method with 96-well microplate; however, this method can be easily realized on glass slide. In this way, this method can be used to realize more high-throughput methylation detection by arraying more capture probes in DNA microarray format. In this case, two advantages would be obtained. One is that the consumption of experimental materials, including oligonucleotides modified with amino groups, primary antibodies, and fluorescently-labeled second antibodies, would be greatly decreased, making this method more cost-effective. The other is that the experimental homogeneity and comparability between various loci would be improved by realizing detecting multiple targets in a very small area. However, this method is suitable for the detection of the methylation levels of interested genomic locations, especially the locations contained the DNA-binding sites of transcription factor.
Conclusions
This study developed a method for DNA methylation detection that based on NIRF technique. The method was validated by detecting methylation of synthesized oligonucleotides and nine genomic loci of the promoters of three genes, KIR3DL1, p14ARF and TP53BP2 in three cancer cell lines. The results were verified by bisulfite sequencing PCR and other previous studies. The method has several advantages over current methods, such as free of bisulfite conversion, PCR amplification and immunoprecipitation. This study thus provides a new tool for the epigenomic studies.
Acknowledgments
Funding: This work was supported by the grants from the National Natural Science Foundation of China (grant number 61171030), Jiangsu Province “The Project of Invigorating Health Care through Science, Technology and Education” special fund, the General Program of the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (grant number 16KJD320005), the Scientific Research Foundation for Excellent Talents of Xuzhou Medical University (grant number D2016014) and Xuzhou Science and Technology Planning Project (grant number KC16SY157).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.01.40). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science 2001;293:1068-70. [Crossref] [PubMed]
- Nadasi E, Clark JS, Szanyi I, et al. Epigenetic modifiers exacerbate oxidative stress in renal proximal tubule cells. Anticancer Res 2009;29:2295-9. [PubMed]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005;6:597-610. [Crossref] [PubMed]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003;349:2042-54. [Crossref] [PubMed]
- Tao L, Wang W, Li L, et al. DNA hypomethylation induced by drinking water disinfection by-products in mouse and rat kidney. Toxicol Sci 2005;87:344-52. [Crossref] [PubMed]
- Ferguson LR, Tatham AL, Lin Z, et al. Epigenetic regulation of gene expression as an anticancer drug target. Curr Cancer Drug Targets 2011;11:199-212. [Crossref] [PubMed]
- Shen L, Waterland RA. Methods of DNA methylation analysis. Curr Opin Clin Nutr Metab Care 2007;10:576-81. [Crossref] [PubMed]
- Hahn MA, Pfeifer GP. Methods for genome-wide analysis of DNA methylation in intestinal tumors. Mutat Res 2010;693:77-83. [Crossref] [PubMed]
- Shiraishi M, Hayatsu H. High-speed conversion of cytosine to uracil in bisulfite genomic sequencing analysis of DNA methylation. DNA Res 2004;11:409-15. [Crossref] [PubMed]
- Bailey VJ, Keeley BP, Razavi CR, et al. DNA methylation detection using MS-qFRET, a quantum dot-based nanoassay. Methods 2010;52:237-41. [Crossref] [PubMed]
- Kinoshita K, Minagawa M, Takatani T, et al. Establishment of diagnosis by bisulfite-treated methylation-specific PCR method and analysis of clinical characteristics of pseudohypoparathyroidism type 1b. Endocr J 2011;58:879-87. [Crossref] [PubMed]
- Yan PS, Wei SH, Huang TH. Methylation-specific oligonucleotide microarray. In: Tollefsbol TO. editor. Epigenetics Protocols. Humana Press, 2004:251-60.
- Kristensen LS, Hansen LL. PCR-based methods for detecting single-locus DNA methylation biomarkers in cancer diagnostics, prognostics, and response to treatment. Clin Chem 2009;55:1471-83. [Crossref] [PubMed]
- Stains CI, Furman JL, Segal DJ, et al. Site-specific detection of DNA methylation utilizing mCpG-SEER. J Am Chem Soc 2006;128:9761-5. [Crossref] [PubMed]
- Tanaka K, Tainaka K, Kamei T, et al. Direct labeling of 5-methylcytosine and its applications. J Am Chem Soc 2007;129:5612-20. [Crossref] [PubMed]
- Hiraoka D, Yoshida W, Abe K, et al. Development of a method to measure DNA methylation levels by using methyl CpG-binding protein and luciferase-fused zinc finger protein. Anal Chem 2012;84:8259-64. [Crossref] [PubMed]
- Yoshida W, Yoshioka H, Bay DH, et al. Detection of DNA Methylation of G-Quadruplex and i-Motif-Forming Sequences by Measuring the Initial Elongation Efficiency of Polymerase Chain Reaction. Anal Chem 2016;88:7101-7. [Crossref] [PubMed]
- Yoshida W, Baba Y, Karube I. Global DNA Methylation Detection System Using MBD-Fused Luciferase Based on Bioluminescence Resonance Energy Transfer Assay. Anal Chem 2016;88:9264-8. [Crossref] [PubMed]
- Vucic EA, Wilson IM, Campbell JM, et al. Methylation analysis by DNA immunoprecipitation (MeDIP). Methods Mol Biol 2009;556:141-53. [Crossref] [PubMed]
- Nair SS, Coolen MW, Stirzaker C, et al. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics 2011;6:34-44. [Crossref] [PubMed]
- Wu H, Coskun V, Tao J, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 2010;329:444-8. [Crossref] [PubMed]
- Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 2005;37:853-62. [Crossref] [PubMed]
- Bock C, Tomazou EM, Brinkman AB, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol 2010;28:1106-14. [Crossref] [PubMed]
- Jacinto FV, Ballestar E, Esteller M. Methyl-DNA immunoprecipitation (MeDIP): hunting down the DNA methylome. Biotechniques 2008;44:35-37, 39 passim. [Crossref] [PubMed]
- Wu M, Wang X, Wang K, et al. Sequence-specific detection of cytosine methylation in DNA via the FRET mechanism between upconversion nanoparticles and gold nanorods. Chem Commun (Camb) 2016;52:8377-80. [Crossref] [PubMed]
- Dadmehr M, Hosseini M, Hosseinkhani S, et al. Label free colorimetric and fluorimetric direct detection of methylated DNA based on silver nanoclusters for cancer early diagnosis. Biosens Bioelectron 2015;73:108-13. [Crossref] [PubMed]
- Kermani HA, Hosseini M, Dadmehr M, et al. Rapid restriction enzyme free detection of DNA methyltransferase activity based on DNA-templated silver nanoclusters. Anal Bioanal Chem 2016;408:4311-8. [Crossref] [PubMed]
- Zhang S, Metelev V, Tabatadze D, et al. Near-infrared fluorescent oligodeoxyribonucleotide reporters for sensing NF-kappaB DNA interactions in vitro. Oligonucleotides 2008;18:235-43. [Crossref] [PubMed]
- Ntziachristos V, Bremer C, Weissleder R. Fluorescence imaging with near-infrared light: new technological advances that enable in vivo molecular imaging. Eur Radiol 2003;13:195-208. [PubMed]
- Hilderbrand SA, Weissleder R. Near-infrared fluorescence: application to in vivo molecular imaging. Curr Opin Chem Biol 2010;14:71-9. [Crossref] [PubMed]
- Chen Y, Pan Y, Zhang B, et al. Analyzing abundance of mRNA molecules with a near-infrared fluorescence technique. Anal Bioanal Chem 2014;406:537-48. [Crossref] [PubMed]
- Zhou F, Xing Y, Xu X, et al. NBPF is a potential DNA-binding transcription factor that is directly regulated by NF-κB. Int J Biochem Cell Biol 2013;45:2479-90. [Crossref] [PubMed]
- Zhou F, Ling X, Yin J, et al. Analyzing transcription factor activity using near infrared fluorescent bridge polymerase chain reaction. Anal Biochem 2014;448:105-12. [PubMed]
- Yin J, Gan P, Zhou F, et al. Sensitive detection of transcription factors using near infrared fluorescent solid-phase rolling circle amplification. Anal Chem 2014;86:2572-9. [Crossref] [PubMed]
- Chen Y, Wang J. A membrane-based near-infrared fluorescence assay for detecting DNA methylation and transcription. Anal Biochem 2013;442:196-204. [Crossref] [PubMed]
- Wang JK, Li JL, Li ML, et al. Assay of DNA-binding proteins with a dsDNA-coupled plate. Clin Biochem 2006;39:167-75. [Crossref] [PubMed]
- Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol 2003;3:235-40. [PubMed]
- Rohde C, Zhang Y, Reinhardt R, et al. BISMA--Fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics 2010;11:230. [Crossref] [PubMed]
- Liu ZJ, Lu X, Zhang Y, et al. Downregulated mRNA expression of ASPP and the hypermethylation of the 5'-untranslated region in cancer cell lines retaining wild-type p53. FEBS Lett 2005;579:1587-90. [Crossref] [PubMed]
- Ito T, Nishida N, Fukuda Y, et al. Alteration of the p14(ARF) gene and p53 status in human hepatocellular carcinomas. J Gastroenterol 2004;39:355-61. [Crossref] [PubMed]
- Zheng S, Chen P, McMillan A, et al. Correlations of partial and extensive methylation at the p14(ARF) locus with reduced mRNA expression in colorectal cancer cell lines and clinicopathological features in primary tumors. Carcinogenesis 2000;21:2057-64. [Crossref] [PubMed]
- Wong JJ, Hawkins NJ, Ward RL. Colorectal cancer: a model for epigenetic tumorigenesis. Gut 2007;56:140-8. [Crossref] [PubMed]
- Gao XN, Yu L. E2F1 contributes to the transcriptional activation of the KIR3DL1 gene. Biochem Biophys Res Comm 2008;370:399-403. [Crossref] [PubMed]
- Wojdacz TK, Møller TH, Thestrup BB, et al. Limitations and advantages of MS-HRM and bisulfite sequencing for single locus methylation studies. Expert Rev Mol Diagn 2010;10:575-80. [Crossref] [PubMed]
- Paz MF, Fraga MF, Avila S, et al. A systematic profile of DNA methylation in human cancer cell lines. Cancer Res 2003;63:1114-21. [PubMed]
- Lind GE, Thorstensen L, Løvig T, et al. A CpG island hypermethylation profile of primary colorectal carcinomas and colon cancer cell lines. Mol Cancer 2004;3:28. [Crossref] [PubMed]
- Esteller M, Cordon-Cardo C, Corn PG, et al. p14ARF silencing by promoter hypermethylation mediates abnormal intracellular localization of MDM2. Cancer Res 2001;61:2816-21. [PubMed]
- Burden AF, Manley NC, Clark AD, et al. Hemimethylation and non-CpG methylation levels in a promoter region of human LINE-1 (L1) repeated elements. J Biol Chem 2005;280:14413-9. [Crossref] [PubMed]
- Vu TH, Li T, Nguyen D, et al. Symmetric and asymmetric DNA methylation in the human IGF2–H19 imprinted region. Genomics 2000;64:132-43. [Crossref] [PubMed]
- Chen D, Padiernos E, Ding F, et al. Apoptosis-stimulating protein of p53-2 (ASPP2/53BP2L) is an E2F target gene. Cell Death Differ 2004;12:358-68. [Crossref] [PubMed]
- Fogal V, Kartasheva NN, Trigiante G, et al. ASPP1 and ASPP2 are new transcriptional targets of E2F. Cell Death Differ 2005;12:369-76. [Crossref] [PubMed]
- Robertson KD, Jones PA. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol 1998;18:6457-73. [Crossref] [PubMed]
- Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc Natl Acad Sci U S A 2000;97:6481-6. [Crossref] [PubMed]


