
Unlocking the epigenetic code of T cell exhaustion
T cell exhaustion is one of the hallmarks of cancer and chronic viral infections. Revitalization of exhausted T cells is critical for restoring immune function and curing diseases. Recent studies suggest that epigenetic mechanisms are directly involved in the transcriptional control of T cell exhaustion. Decoding the epigenome specific to exhausted T cells is a pivotal step toward development of therapeutic approaches for overcoming T cell exhaustion in infections and cancer.
In chronic infections and cancer, patients’ T cell function gradually deteriorates over time due to persistent exposure to antigens and/or inflammatory stimuli, resulting in a state called T cell “exhaustion” (1). Exhausted T cells are generally associated with poor effector function, loss of proliferative capacity, impaired cytotoxicity, reduced cytokine production, and inefficient control of infections and tumors. The hallmarks of exhausted T cells include overexpression of multiple inhibitory receptors such as PD-1 (CD279), cytotoxic T lymphocyte antigen-4 (CTLA-4, CD152), lymphocyte-activation gene 3 (Lag-3), T cell immunoglobulin domain 3 (Tim-3), CD244/2B4, CD160, and T cell immunoreceptor with Ig and ITIM domain (TIGIT), and an impaired ability to produce cytokines such as IFN-γ, TNF-α, and IL-2 (1). These disarmed T cells sit on the sidelines of the battlefield of immune systems, being fettered by the underlying molecular inhibitory mechanisms, and thus succumb to the growth of intruders such as viruses and tumors. The discovery that immune checkpoint blockade can rejuvenate the exhausted T cells and lead to reduced tumor load was truly a breakthrough in the cancer immunology and immunotherapy field (2). However, questions concerning the persistence of effectiveness still need to be addressed even though T cell function appears to be restored and improved in the presence of checkpoint inhibitors (i.e., PD-1/PD-L1 inhibitors). Chimeric antigen receptor (CAR) T cell therapy has emerged as a promising immunotherapy approach for hematologic malignancy and achieved curative effects in some cases; however, the application of CAR T cell therapy in solid tumors is still quite challenging. The induction of exhaustion in the engineered T cells by persistent CAR signaling remains a substantial barrier to overcome (3). Thus, it is necessary to understand the mechanisms that regulate exhausted T cells with an ultimate goal of developing more effective strategies for immunotherapy.
Growing evidence suggests that epigenetic regulation (i.e., DNA methylation and histone modifications) plays an important role in the differentiation of T cells (4). The switch to an “exhaustion” phenotype in T cells is most likely the result of an altered epigenome that controls global transcription program. For examples, during CD8+ T cells differentiation in the context of acute viral infections, the regulatory region of Pdcd1, which encodes mouse PD-1, became transiently demethylated in the effector phase, but remethylated in functional memory CD8+ T cells as infections resolved (5). By contrast, as illustrated in Figure 1, the same Pdcd1 locus was completely demethylated in exhausted CD8+ T cells from chronic infections and remained unmethylated even when virus titers and PD-1 protein expression by exhausted CD8+ T cells decreased (5). This lack of DNA remethylation leaves the Pdcd1 gene poised for rapid expression, potentially providing a signal for premature termination of antiviral functions. It has also been observed in CD8+ T cells from leukemia patients that the PD-1 expression level was augmented in accordance with a decrease in DNA methylation levels in a distal regulatory region of human PDCD1 (7). Moving beyond DNA methylation, one of the extensively studied epigenetic marks, Austin and colleagues (8) identified a number of novel distal regulatory regions within the Pdcd1 locus based on chromatin accessibility and demonstrated that induction and sustained expression of PD-1 is regulated at the chromatin level via long-range cis-elements interactions in murine T cells.
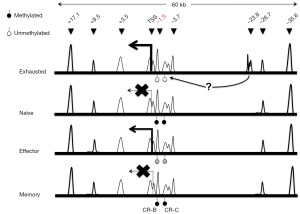
Most recently, to further identify epigenetic regulatory regions in the genome of exhausted T cells, Sen et al. mapped the chromatin accessible regions (ChARs) in mouse T cells infected with lymphocytic choriomeninigits virus (LCMV) (6) using a recently developed technology called ATAC-seq, which stands for Assay for Transposase-Accessible Chromatin with high-throughput sequencing. ATAC-seq is a more sensitive and simple alternative to DNase-Seq for assaying genome-wide chromatin accessibility. It uses Tn5 transposase to probe DNA accessibility, followed by next-generation sequencing to identify regions with gain or loss of chromatin accessibility across the genome (9). Sen et al. analyzed high quality ATAC-seq data and revealed that chromatin accessibility appeared to be increased throughout the mouse genome during naïve CD8+ T cell differentiation (6). Compared with functional effector or memory CD8+ T cells, exhausted T cells exhibited a significant reorganization of ChARs in regulatory regions, which are noticeably enriched at intergenic elements and introns. Moreover, the average peak intensity of ChARs positively correlated with the average expression of their adjacent genes, suggesting presence of potential enhancer elements within ChARs. Among those clustered ChARs assembled in modules by similarity, some modules and adjacent genes were explicitly present during CD8+ T cell differentiation. Interestingly, the magnitude of difference in the profile of ChARs between exhausted and functional CD8+ T cells was far greater than that seen in gene expression profiles. Furthermore, exhausted CD8+ T cells contained ChAR modules that were associated with increased expression of inhibitory receptors Pdcd1 and Havcr2 and the transcriptional factor Batf, as well as the pathways involved in negative regulation of cytokine production and T cell activation. These results further stress the importance of epigenetic regulation in the processes leading to T cell exhaustion.
Next, researchers focused on the inhibitory receptor PD-1 to gain further insights into the role that specific chromatin accessible regions played in regulating genes differentially expressed in exhausted CD8+ T cells. In addition to the previously known Pdcd1 regulatory regions in CD8+ T cells from both acute and chronic infections, ATAC-seq analysis identified a ChAR in an upstream region of Pdcd1 (−23.8 kb) that was only observed in exhausted CD8+ T cells from chronic LCMV infections (Figure 1). This region contains enhancer elements as revealed by reporter assays; and its necessity for sustaining high level PD-1 expression was confirmed using CRISPR/Cas9-based deletion of a 1.2-kb fragment within the −23.8 kb enhancer region in mouse EL4 cells. The highest amount of enhancer deletion was observed in cells with lowest PD-1 expression, indicating the importance of this 1.2-kb regulatory element. To dissect the functional contribution of specific sequences within each enhancer element, Sen and colleagues employed a CRISPR/Cas9-based screening approach with a pool of 1,754 sgRNAs targeting the −23.8 kb enhancer and eight other regulatory regions and exons. By comparing the abundance of each sgRNA between sorted PD-1- high and PD-1- low EL4 cells co-transduced with Cas9 and the sgRNA library, the research team showed that eight out of nine regulatory regions investigated contained sequences that were critical for maintaining PD-1 expression in EL4 cells. When focusing on the −23.8 kb enhancer, three sgRNA cleavage-sites were found to be associated with remarkable down-regulation of PD-1 expression. Interestingly, analysis of putative transcription factor (TF) binding sites near the three cleavage-sites revealed specific enrichment of Sox3, T-bet and retinoic acid receptor (RAR) motifs.
Overall, Sen et al. showed that 80–85% of ChARs defined in the mouse model could be mapped to their human orthologous regions; Similar to mouse models, human CD8+ T cells differentiated over time were accompanied by changes in chromatin accessibility, permitting enhancer activity and TF binding (6). However, the −23.8 kb Pdcd1 enhancer region was not among those mapped from the mouse model, suggesting that a similar analysis of the epigenome in exhausted human T cells is necessary for better understanding of T cell exhaustion in the context of human diseases.
Genome-wide sequencing analysis at chromatin levels has brought to light novel understanding of the connection between a gene and its epigenetic co-regulatory elements. Chromatin accessible regions can be reprogrammed to open up to TF binding, chromatin remodeling and subsequent activation of gene expression. Moreover, recent studies provided a 3D view into the topological organization of chromatin structure, which allows an orchestration of gene expression through long-range interactions of cis-regulatory elements in various types of cells (10). Exhausted CD8+ T cells possess a distinct functional state encoded by the concomitant chromatin accessibility through the activation of exhaustion-specific cis-regulatory elements. Mapping of these state-specific cis-regulatory regions such as enhancers in exhausted CD8+ T cells may lead to more precise genome editing for adoptive T cell therapy (6). CRISPR/Cas9-based gene-editing of CAR-T cells to make them resistant to exhaustion by deleting the PD-1 gene locus has been implemented in the first human trial recently (11). As CRISPR/Cas9-based epigenetic targeting approaches become more practical, it will soon become a reality for targeted site-specific modification of epigenetic marks such as DNA methylation and histone modification in human cells without physically deleting DNA (12-14). In the future, reversing T cell exhaustion may be accomplished by inactivating exhaustion-specific enhancers via epigenome editing approaches, which may provide a more “tunable” and state-specific approach to modulate T cell function than deleting the target genes. Further studies are warranted to define the global epigenetic changes in exhausted T cells in the context of various human diseases and develop more faithful epigenome editing technologies. Such advances will bring the re-armed T cells back to the immune battleground and pave the way for successful therapeutic opportunities.
Acknowledgments
Funding: This work was supported in part by the National Institute of Health (Grants CA185833 and GM113242 to H.S.). H.S. is a Georgia Cancer Coalition Distinguished Cancer Scientist.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Yi Miao (Department of Hematology, The First Affiliated Hospital of Nanjing Medical University, Jiangsu Province Hospital, Nanjing, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.03.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015;15:486-99. [Crossref] [PubMed]
- Xia Y, Medeiros LJ, Young KH. Immune checkpoint blockade: Releasing the brake towards hematological malignancies. Blood Rev 2016;30:189-200. [Crossref] [PubMed]
- Long AH, Haso WM, Shern JF, et al. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat Med 2015;21:581-90. [Crossref] [PubMed]
- Youngblood B, Hale JS, Ahmed R. T-cell memory differentiation: insights from transcriptional signatures and epigenetics. Immunology 2013;139:277-84. [Crossref] [PubMed]
- Youngblood B, Oestreich KJ, Ha SJ, et al. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity 2011;35:400-12. [Crossref] [PubMed]
- Sen DR, Kaminski J, Barnitz RA, et al. The epigenetic landscape of T cell exhaustion. Science 2016;354:1165-9. [Crossref] [PubMed]
- Wu J, Xu X, Lee EJ, et al. Phenotypic alteration of CD8+ T cells in chronic lymphocytic leukemia is associated with epigenetic reprogramming. Oncotarget 2016;7:40558-70. [PubMed]
- Austin JW, Lu P, Majumder P, et al. STAT3, STAT4, NFATc1, and CTCF regulate PD-1 through multiple novel regulatory regions in murine T cells. J Immunol 2014;192:4876-86. [Crossref] [PubMed]
- Buenrostro JD, Giresi PG, Zaba LC, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 2013;10:1213-8. [Crossref] [PubMed]
- Achinger-Kawecka J, Clark SJ. Disruption of the 3D cancer genome blueprint. Epigenomics 2017;9:47-55. [Crossref] [PubMed]
- Cyranoski D. CRISPR gene-editing tested in a person for the first time. Nature 2016;539:479. [Crossref] [PubMed]
- Hilton IB, D'Ippolito AM, Vockley CM, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 2015;33:510-7. [Crossref] [PubMed]
- Liu XS, Wu H, Ji X, et al. Editing DNA Methylation in the Mammalian Genome. Cell 2016;167:233-47.e17. [Crossref] [PubMed]
- Xu X, Tao Y, Gao X, et al. A CRISPR-based approach for targeted DNA demethylation. Cell Discov 2016;2:16009. [Crossref] [PubMed]


