
Are all ALK rearrangements created equal?
The discovery of ALK rearrangements in non-small cell lung cancer (NSCLC) and their oncogenic properties by Soda et al. in 2007 gave the start to one of the most famous stories in the treatment of lung cancer (1). Diagnosed in around 5% of patients with NSCLC, ALK rearrangements are effectively treated with the ALK-directed tyrosine kinase inhibitor (ALK-TKI) crizotinib (2,3). Two phase III trials have demonstrated that crizotinib is superior to standard chemotherapy in first- and second-line settings, the objective response rates ranging from 65% to 74% with median progression-free survivals (PFS) of 7.7 to 10.9 months, and crizotinib is thus currently the standard of care in advanced ALK-positive NSCLC, in first-line setting (3). This justifies the search for ALK-rearrangements in all advanced non-squamous NSCLCs, especially in adenocarcinomas of cribiform/signet-ring cells subtypes, non-smoker and young patients (4). Identification of ALK-positive NSCLCs is based on immunohistochemistry (antibodies 5A4 Novocastra™ and D5F3 Cell Signaling™), break-apart fluorescent in situ hybridization (FISH), fusion variants specific multiplex RT-PCR, Nanostring nCounter® technology and targeted RNA-sequencing assays (5,6). By these latter techniques several variants can be identified for which prognostic or predictive values remain to be elucidated.
Nevertheless, individual responses and PFS observed with crizotinib in clinical trials and in routine practice are heterogeneous (3,7). Some patients experience a progressive disease as best response, others relapse within a few months after crizotinib initiation, and the last remain with persistent objective responses after years of treatment. The inability to achieve a therapeutic concentration of crizotinib in the central nervous system is probably the main mechanism of early brain oligometastatic progression observed in some patients (3). On the other hand, the molecular mechanisms underlying early diffuse progression are poorly understood, even if uncommon false positive ALK diagnosis, lack of observance and pharmacological interactions may be in cause for a few patients (3). One of the main hypotheses that have been raised is based on the different protein stabilities of EML4-ALK fusion variants products. Indeed, various parts of EML4 that are fused to ALK in the different variants influence the fusion proteins stability, inhibitor-induced protein degradation, and drug sensitivity. Heuckmann et al. induced the expression of four different EML4-ALK fusion variants (v1, v2, v3a and v3b) in a BaF3 cell line model and showed that the cell line sensitivity to crizotinib and the tool compound ALK-TKI TAE684 correlated with the expected protein stability of the EML4-ALK variants (8). EML4-ALK v2 was the most sensitive to ALK-TKI, EML4-ALK v1 and v3b had intermediate sensitivity and V3a was less sensitive. Until the recent article of Woo et al. published in Annals of Oncology accompanying this editorial, these in vitro data suggesting that ALK rearrangements are not created equal had never been corroborated by clinical observations (9).
The authors report the results of a single institution, retrospective analysis of crizotinib efficacy according to EML4-ALK fusion variants among a population of 54 Korean patients with ALK-positive advanced NSCLC. Using a multiplex RT-PCR, the authors identified 28 known EML4-ALK variants in tumor samples. An EML4-ALK v3a/b was identified in 24 cases (44.4%), a v1 in 18 (33.3%) and a v2 in 6 (11.1%). Rare ALK transcripts were detected in the 6 remaining cases (v7 in one, two new EML4-ALK variants in two and non-EML4 variants in three). According to the expected greater instability of EML4-ALK v1 and v2 compared to v3a/b, Woo et al. cleverly compared ALK-TKI efficacy between a v1/v2/others group (n=27) and a v3a/b group (n=24), excluding from the analysis the non-EML4-ALK variants. In the 44 patients treated with crizotinib, the 2-year PFS rate was clearly improved in the v1/v2/others group compared to the v3a/b group (76.0% vs. 26.4%, P=0.034). After adding the seven patients treated with the second-generation ALK-TKIs ceritinib or alectinib, the 2-year PFS rate was numerically improved in the v1/v2/others group compared to the v3a/b group but this did not reach statistical significance (69.0% vs. 32.7%, P=0.108). Objective response rates and disease control rates were slightly superior in the v1/v2/others group, especially when considering all ALK-TKIs, but with no statistical significance. In IL-3-dependent Ba/F3 cells that stably expressed v1, v2, v3a or v5a, ALK tyrosine kinase activity was shown to be higher in v3a- and V5a-expressing cells. The v3a- and v5a-expressing cells were also resistant to crizotinib, ceritinib and alectinib (IC50 >500nM), as were v3a-expressing H2228 and v5a-expressing BEAS-2B cell lines, whereas v1- and v2-expressing cells were sensitive to ALK-TKIs.
To date, this is the first study that supports the notion that EML4-ALK variants are able to condition the clinical benefit of ALK-TKIs according to the different stabilities of the protein products in ALK-positive NSCLC. However, apparent conflicting data have emerged from the literature (Table 1). Indeed, a single-institution retrospective study of 35 ALK-positive patients recently published by Yoshida et al. suggested that crizotinib was more effective in EML4-ALK v1 versus non-v1 variants (12). Apart from the small size of the cohort studied, the comparison of EML4-ALK v1 versus non-v1 was not supported by any biological rationale. A careful analysis of the individual PFS provided in this paper shows that the PFS was always inferior to 5 months in the four EML4-ALK v3a/b patients, which is indeed lower than what is expected with crizotinib in ALK-positive NSCLC. Another retrospective study in 61 ALK-positive patients showed lower response rates in EML4-ALK v3a/b patients compared to EML4-ALK v1 but no differences in PFS were observed (11). Finally, a retrospective study reported clinical outcomes with crizotinib in 52 ALK-positive patients and showed no differences in response rates between EML4-ALK v1 and EML4-ALK v3a/b, but a better PFS in EML4-ALK v1 compared to EML4-ALK v3a/b (13). Considering these scarce data available in the literature, the findings from the paper of Woo et al., together with the work from Heuckmann et al., seem to be relevant. Are these new data strong enough to suggest that ALK rearrangements should not be considered equal in the clinic?

Full table
Some important limitations have to be underlined in this work (9). First, a selection bias is suspected that is inherent to the retrospective nature of this study. From the 182 patients initially diagnosed with an ALK-rearranged NSCLC, only 54 patients were finally analyzed. Importantly, 81 samples from patients treated with ALK-TKI were available and among them, 24 were excluded because of RT-PCR failure. In numerous studies that report the frequency distribution of EML4-ALK variants, the v1 is the more common (around 50% of the cases), followed by v3a/b (around 25%) and v2 (around 10%) (3). The distribution reported herein shows a predominance of v3a/b (44.4%) followed by v1 (33.3%) and v2 (11.1%). This probably does not reflect the distribution in the overall ALK-positive NSCLC population and therefore questions the representativity of the cohort that has been analyzed.
Second, the 2-year PFS rate was improved in the v1/v2/others group compared to the v3a/b group when considering the 44 patients treated with crizotinib. This difference did not translate into overall survival (OS), with stackable curves in the two groups. The authors argued that this could be due to the low mortality rate and a larger proportion of patients treated with front-line ALK-TKI in the v3a/b group. These assumptions are right but can only partially explain the lack of OS differences. The progression was defined according to RECIST criteria, as recommended in clinical trials. However, it is very well known that RECIST-defined progression is not always a marker of treatment failure in the field of oncogene-addiction (14). In ALK-positive NSCLC, oligoprogression as first progression event is not uncommon and could be effectively treated with local ablative therapy and continuing ALK-TKI (15). Continuing crizotinib beyond progression is also a widely used strategy when a clinical benefit of the treatment is still observed (7,16). ALK-positive NSCLC patients that could be managed by this “treatment beyond progression” exhibit a very good prognosis (7,16). As no data are shown about the profile of progression, it cannot be excluded that this subgroup of ALK-positive NSCLC with oligoprogressive disease and/or treated with crizotinib beyond progressive disease is over-represented in the v3a/b group, therefore explaining the lack of OS differences. Furthermore, no information was provided about next-generation ALK-TKI that some patients probably received after crizotinib failure. Next-generation ALK-TKIs are effective in this setting, and imbalance of next-generation ALK-TKI treatment after crizotinib failure favoring the v3a/b group could also explain the lack of OS differences (3). The 2-year PFS rate improvement in the v1/v2/others group was no longer observed when the seven patients treated with next-generation ALK-TKI ceritinib and alectinib were added to the analysis, rising the hypothesis that the efficacy of these ALK inhibitors are less impacted by the EML4-ALK different variants. As ceritinib and alectinib have a better ALK tyrosine kinase inhibitor activity than crizotinib, one could postulate that these former drugs inhibit more effectively v3a/b ALK variants despite a more stable protein product. However, in vitro experiments reported by Woo et al. did not support this hypothesis. Interestingly, preliminary data from ASCEND-1 clinical trial testing ceritinib after crizotinib failure or in ALK-TKI naïve-patients also suggest that PFS with ceritinib is higher in v1 patients than in v3 patients (17). More data are needed to elucidate whether next-generation ALK-TKI affect differently EML4-ALK variants, or not.
Third, the multiplex RT-PCR assay used to detect the EML4-ALK variants is not able to distinguish the variant 3a from the variant 3b. In the original study that suggested the role of different EML4-ALK variants stability in crizotinib efficacy, v2 was the most sensitive, v1 and v3b had intermediate sensitivities and 3a was the least sensitive to crizotinib (8). Pooling v1 and v2 variants then v3a and v3b variants in the same group could be considered somewhat artificial in regard of these previous results. However, experiment correlates from the paper of Woo et al. suggest that v1-expressing BEAS-2B cells were more sensitive to crizotinib than v2- and v3a-variants (9). Unfortunately, no v3b-expressing BEAS-2B was generated but the H2228 cell line which contains a v3b variant was resistant to crizotinib. These results are in line with the clinical correlates but conflicting with the original study from Heuckmann et al. (8).
Non-EML4-ALK rearrangements are not detectable with the assay used by Woo et al. To date, at least 21 non-EML4 partner genes have been described in NSCLC and very few data about sensitivity to ALK-TKI in the clinic are available (Table 2) (1,10,17-30). Partial responses to crizotinib are commonly reported in clinical cases but PFS are very heterogeneous, ranging from 6 months to 28 months (Table 2). In RET-rearranged NSCLC, differences in vandetanib RET-TKI efficacy has been shown according to RET rearrangement variants (31). Responses and PFS were more favorable in CCDC6-RET than in KIF5B-RET fusion. Despite the fact that these kinds of rearrangements are probably uncommon, more data are needed to evaluate the impact of non-EML4-ALK rearrangements on ALK-TKI efficacy.
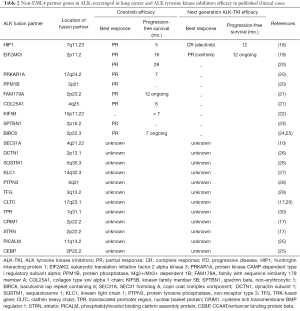
Full table
Finally, the findings of Woo et al. raise the question whether all ALK rearrangements are created equal. Considering the conflicting results in the literature and the limitations that have been highlighted herein, these data are too preliminary to answer this question and impact our treatment decision in ALK-rearranged NSCLC. However, this hypothesis should be better elucidated using NGS technology to explore all variants, including non-EML4-ALK variants, in a more scalable way than multiplex RT-PCR. This NGS diagnostic approach should be considered in clinical trials to enhance the understanding of ALK variants biology, clinical course and impact in treatment efficacy and make a step forward in its clinical application as a relevant predictive biomarker.
Acknowledgments
Funding: MD has received research funding from Novartis and Pfizer. He has been reimbursed for travel, accommodation, and/or other expenses by Novartis, Pfizer and Roche; DMS has received research funding from Pfizer; JC has received research funding from Novartis and Pfizer.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Shaohua Cui (Department of Pulmonary Medicine, Shanghai Chest Hospital, Shanghai Jiaotong University, Shanghai, China).
Conflicts of Interest: MD has received research funding from Novartis and Pfizer. He has been reimbursed for travel, accommodation, and/or other expenses by Novartis, Pfizer and Roche; AMF has declared no conflicts of interest; DMS has received research funding from Pfizer. He has served as a consultant (advisory board) for Novartis, Pfizer and Roche; JC has received research funding from Novartis and Pfizer. He has served as a consultant (advisory board) for Novartis, Pfizer and Roche.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Barlesi F, Mazieres J, Merlio JP, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 2016;387:1415-26. [Crossref] [PubMed]
- Lin JJ, Riely GJ, Shaw AT. Targeting ALK: Precision Medicine Takes on Drug Resistance. Cancer Discov 2017;7:137-55. [Crossref] [PubMed]
- Fallet V, Cadranel J, Doubre H, et al. Prospective screening for ALK: clinical features and outcome according to ALK status. Eur J Cancer 2014;50:1239-46. [Crossref] [PubMed]
- Lantuejoul S, Rouquette I, Blons H, et al. French multicentric validation of ALK rearrangement diagnostic in 547 lung adenocarcinomas. Eur Respir J 2015;46:207-18. [Crossref] [PubMed]
- Heydt C, Kostenko A, Merkelbach-Bruse S, et al. ALK evaluation in the world of multiplex testing: Network Genomic Medicine (NGM): the Cologne model for implementing personalised oncology. Ann Oncol 2016;27:iii25-34. [Crossref] [PubMed]
- Duruisseaux M, Besse B, Cadranel J, et al. Overall survival with crizotinib and next-generation ALK inhibitors in ALK-positive non-small-cell lung cancer (IFCT-1302 CLINALK): a French nationwide cohort retrospective study. Oncotarget 2017; [Epub ahead of print]. [Crossref]
- Heuckmann JM, Balke-Want H, Malchers F, et al. Differential protein stability and ALK inhibitor sensitivity of EML4-ALK fusion variants. Clin Cancer Res 2012;18:4682-90. [Crossref] [PubMed]
- Woo CG, Seo S, Kim SW, et al. Differential protein stability and clinical responses of EML4-ALKfusion variants to various ALK inhibitors in advanced ALK-rearranged non-small cell lung cancer. Ann Oncol 2016; [Epub ahead of print]. [Crossref] [PubMed]
- Kim RN, Choi YL, Lee MS, et al. SEC31A-ALK Fusion Gene in Lung Adenocarcinoma. Cancer Res Treat 2016;48:398-402. [Crossref] [PubMed]
- Lei YY, Yang JJ, Zhang XC, et al. Anaplastic Lymphoma Kinase Variants and the Percentage of ALK-Positive Tumor Cells and the Efficacy of Crizotinib in Advanced NSCLC. Clin Lung Cancer 2016;17:223-31. [Crossref] [PubMed]
- Yoshida T, Oya Y, Tanaka K, et al. Differential Crizotinib Response Duration Among ALK Fusion Variants in ALK-Positive Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3383-9. [Crossref] [PubMed]
- Cha YJ, Kim HR, Shim HS. Clinical outcomes in ALK-rearranged lung adenocarcinomas according to ALK fusion variants. J Transl Med 2016;14:296. [Crossref] [PubMed]
- Oxnard GR, Morris MJ, Hodi FS, et al. When progressive disease does not mean treatment failure: reconsidering the criteria for progression. J Natl Cancer Inst 2012;104:1534-41. [Crossref] [PubMed]
- Weickhardt AJ, Scheier B, Burke JM, et al. Local ablative therapy of oligoprogressive disease prolongs disease control by tyrosine kinase inhibitors in oncogene-addicted non-small-cell lung cancer. J Thorac Oncol 2012;7:1807-14. [Crossref] [PubMed]
- Ou SH, Jänne PA, Bartlett CH, et al. Clinical benefit of continuing ALK inhibition with crizotinib beyond initial disease progression in patients with advanced ALK-positive NSCLC. Ann Oncol 2014;25:415-22. [Crossref] [PubMed]
- Tan DS, Kim DW, Thomas M, et al. Genetic landscape of ALK+ non-small cell lung cancer (NSCLC) patients (pts) and response to ceritinib in ASCEND-1. J Clin Oncol 2016;34 suppl:abstr 9064.
- Ou SH, Klempner SJ, Greenbowe JR, et al. Identification of a novel HIP1-ALK fusion variant in Non-Small-Cell Lung Cancer (NSCLC) and discovery of ALK I1171 (I1171N/S) mutations in two ALK-rearranged NSCLC patients with resistance to Alectinib. J Thorac Oncol 2014;9:1821-5. [Crossref] [PubMed]
- Won B, Mambetsariev I, Salgia R. Post-crizotinib management of effective ceritinib therapy in a patient with ALK-positive non-small cell lung cancer. BMC Cancer 2016;16:568. [Crossref] [PubMed]
- Ali SM, Hensing T, Schrock AB, et al. Comprehensive Genomic Profiling Identifies a Subset of Crizotinib-Responsive ALK-Rearranged Non-Small Cell Lung Cancer Not Detected by Fluorescence In Situ Hybridization. The Oncologist 2016;21:762-70. [Crossref] [PubMed]
- Cui S, Zhang W, Xiong L, et al. Use of capture-based next-generation sequencing to detect ALK fusion in plasma cell-free DNA of patients with non-small-cell lung cancer. Oncotarget 2017;8:2771-80. [PubMed]
- Takeuchi K, Choi YL, Togashi Y, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res 2009;15:3143-9. [Crossref] [PubMed]
- Gu FF, Zhang Y, Liu YY, et al. Lung adenocarcinoma harboring concomitant SPTBN1-ALK fusion, c-Met overexpression, and HER-2 amplification with inherent resistance to crizotinib, chemotherapy, and radiotherapy. J Hematol Oncol 2016;9:66. [Crossref] [PubMed]
- Shan L, Jiang P, Xu F, et al. BIRC6-ALK, a Novel Fusion Gene in ALK Break-Apart FISH-Negative Lung Adenocarcinoma, Responds to Crizotinib. J Thorac Oncol 2015;10:e37-39. [Crossref] [PubMed]
- Li W, Zhang J, Guo L, et al. Combinational Analysis of FISH and Immunohistochemistry Reveals Rare Genomic Events in ALK Fusion Patterns in NSCLC that Responds to Crizotinib Treatment. J Thorac Oncol 2017;12:94-101. [Crossref] [PubMed]
- Iyevleva AG, Raskin GA, Tiurin VI, et al. Novel ALK fusion partners in lung cancer. Cancer Lett 2015;362:116-21. [Crossref] [PubMed]
- Togashi Y, Soda M, Sakata S, et al. KLC1-ALK: a novel fusion in lung cancer identified using a formalin-fixed paraffin-embedded tissue only. PloS One 2012;7:e31323 [Crossref] [PubMed]
- Jung Y, Kim P, Jung Y, et al. Discovery of ALK-PTPN3 gene fusion from human non-small cell lung carcinoma cell line using next generation RNA sequencing. Genes Chromosomes Cancer 2012;51:590-7. [Crossref] [PubMed]
- Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190-203. [Crossref] [PubMed]
- Choi YL, Lira ME, Hong M, et al. A novel fusion of TPR and ALK in lung adenocarcinoma. J Thorac Oncol 2014;9:563-6. [Crossref] [PubMed]
- Yoh K, Seto T, Satouchi M, et al. Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): an open-label, multicentre phase 2 trial. Lancet Respir Med 2017;5:42-50. [Crossref] [PubMed]


