The downregulation of NCRUPAR is associated with the clinical characteristics of hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third most common cause of cancer-related deaths worldwide. All the cases (and deaths), 82% are in developing countries and the death rate is the second highest in China because of the high prevalence of chronic hepatitis B virus infection and liver cirrhosis (1,2). HCC usually remains asymptomatic in the early stage. When symptomatic, patients are often at an advanced stage and have lost the opportunity to undergo radical surgery. Although there have been many advances in HCC therapy, such as targeted therapies, liver transplantation, proton therapy, and interventional radiology, the overall patient outcome has not been improved substantially and the 5-year survival rate is 25–39% after surgery (3-8). In addition to the classical histological and molecular markers, the identification of novel HCC targets will enhance our understanding of cancer heterogeneity and facilitate improvement of personalized therapeutic strategies (9). Therefore, it is important to identify a reliable clinical marker for HCC diagnosis and prediction of the clinical outcome.
Long noncoding RNAs (lncRNAs) are novel RNA molecules greater than 200 nucleotides in length that are not translated into proteins. Although these long non-coding transcripts were once considered to be simply transcriptional “noise” or cloning artifacts, recent evidence shows that lncRNAs play important roles in diverse biological processes, such as transcriptional regulation, cell growth, and tumorigenesis (10-14). For example, the overexpression of HOTAIR is associated with poor prognosis in liver (15) and gastrointestinal (16), cancers. A non-protein-coding RNA, UCA1, also increased tumorigenic potential of bladder cancer (17). There is also a relationship between deregulated lncRNAs and HCC (18-20). Unfortunately, the functional role of lncRNAs in HCC remains largely unknown. Recently, a novel lncRNA upstream from the coagulation factor II thrombin receptor (F2R/PAR1) gene, termed NCRUPAR, was identified. NCRUPAR upregulates PAR-1 expression during embryonic growth and might inhibit tumor progression in gastric and colorectal cancer (21-23). However, the clinical and prognostic significance of NCRUPAR expression in HCC has not been reported.
Therefore, this study investigated the expression of NCRUPAR in HCC specimens and adjacent normal tissues and the potential relationship between NCRUPAR expression levels and the clinicopathological factors of patients with HCC. Our data suggest NCRUPAR may be a potential novel biomarker for HCC.
Methods
Specimens and clinical data collection
Our cancer center in the Department of Hepato-biliary and Pancreatic Surgery, The First Affiliated Hospital, College of Medicine, Zhejiang University, China, collected 137 samples of HCC tissues compared with paired adjacent non-tumor tissues from 2010 to 2013. Tumor tissues and paired adjacent non-tumorous tissues 5 cm from the edge of the tumor were obtained during surgery. None of the patients received preoperative chemotherapy or radiation therapy. All tissues were preserved in RNA fixer (BioTeke, Beijing, China) at −80 °C until use. The diagnosis of each specimen was confirmed histopathologically. All clinical data were collected by physicians and the researchers were blinded to the clinical data. The study was approved by the Human Research Ethics Committee of The First Affiliated Hospital, College of Medicine, Zhejiang University. Written informed consent was obtained from all subjects.
Total RNA preparation and qRT-PCR detection
The TRIzol reagent (Invitrogen) was used to extract total RNA from cell lines or tissues according to the man-ufacture’s instructions. The concentration and purity of RNA were controlled by UV spectrophotometry (A260/A280 >1.9) using a nanophotometer UV/Vis spectrophotometer. cDNAs from all samples were synthesized from 500 ng of total RNA using iScript™ cDNA Synthesis Kit (Bio-Rad) following the manufacturer’s protocol. The 20 µ RT reactions were incubated for 5 min at 25 degree centigrade and for 30 min at 42 degree centigrade and were then for 5 min at degree centigrade, in the end maintained at 4 degree centigrade.
The expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and NCRUPAR were evaluated using real-time qRT-PCR. The primers were as follows: NCRUPAR forward (5'-GAGGCAGTAGAATGGCGTAAACC-3) and reverse (5'-TCTAATGCCCGTCTTTTTGCTC-3); and GAPDH forward (5'-AGAAGGCTGGGGCTCATTTG-3) and reverse (5'-AGGGGCCATCCACAGTCTTC-3) (23). The cDNAs were amplified using an Applied Biosystems 7500-fast PCR machine; the reaction was finished according to the PCR kit instructions. Denaturation was at 94 °C for 5 min and followed by 40 cycles of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 20s. All experiments were conducted three times and the average was determined. The formula 2−ΔΔCt was used to calculate the differential gene expression (23).
Cell lines and cell culture
Six HCC cell lines (HepG2, HCCLM3, HUH7, MHCC97H, SK-Hep1 and Hep3B) and one normal liver cell line (QSG-7701), all of which are maintained at our institution, were used in this study. All cells were maintained in a humidified atmosphere containing 5% CO2 at 37 °C and were passaged using standard cell culture techniques.
Statistical analysis
All statistical analyses were performed using SPSS 19.0 for Windows (SPSS, Chicago, IL, USA) and GraphPad Prism5.0 (GraphPad Software, La Jolla, CA, USA). One-way analysis of variance (ANOVA) and Student’s t-test were used as appropriate. The relationship between NCRUPAR expression and the clinicopathological variables was assessed using the χ2 test. The Kaplan-Meier method estimating the 5-year recurrence free survival rates. Statistical significance was accepted at P<0.05.
Results
Expression of NCRUPAR in HCC cell lines and tissues
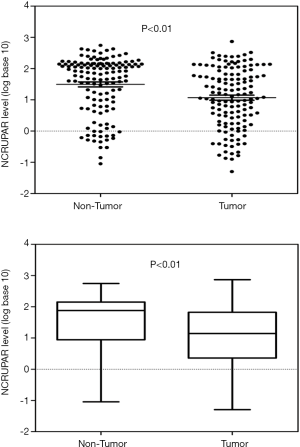
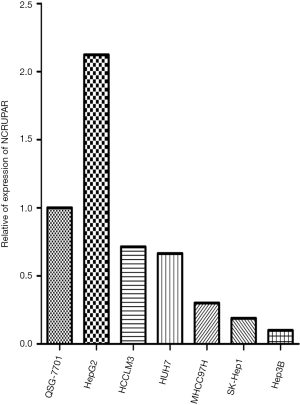
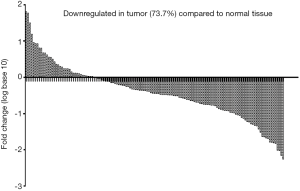
To ascertain whether NCRUPAR expression was also reduced in HCC, as in colorectal and gastric cancer, we first examined the NCRUPAR expression level in HCC cell lines using quantitative reverse transcription (qRT)-PCR and found that the expression of NCRUPAR in five HCC cell lines, HCCLM3, HUH7, MHCC97H, SK-Hep1 and Hep3B, was significantly downregulated compared to a normal liver cell line QSG-7701 (Figure 1). Based on these findings, we further quantified NCRUPAR in paired primary cancerous and adjacent non-cancerous tissues from 137 HCC patients. one hundred and one cases (73.7%) showed significant downregulation of NCRUPAR in cancerous tissues (Figure 2). In all 137 tissue pairs, the NCRUPAR expression was significantly lower in tumors compared to adjacent non-cancerous tissues (P<0.01; Figure 3).
Relationship between NCRUPAR expression and the clinicopathological features of HCC
The relationship with the clinical characteristics was analyzed and the results are shown in Table 1. The NCRUPAR level was associated with cancer distal metastasis (P=0.046), portal vein tumor thrombus (P=0.046) and histopathological grade (P=0.006) However, there was no significant correlation between NCRUPAR expression and other clinicopathological features, such as age, gender, tumor diameter, hepatitis B, liver cirrhosis, clinical stage, or number of tumors and so on.
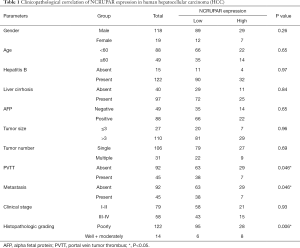
Full table
The survival outcome of HCC Patients according to the level of expression of the NCRUPAR in patients with HCC
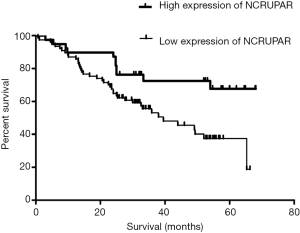
The Kaplan-Meier curve revealed that patients with low expression of NCRUPAR has a worse overall survival compared to patient with high expression of NCRUPAR (P<0.05 Figure 4).
Discussion
The conventional view of gene regulation in biology has centered on protein-coding genes via the central dogma DNA → mRNA → protein. Although initially thought to be spurious transcriptional noise, the discovery of thousands of long non-coding RNAs (lncRNAs) has changed our view of the complexity of mammalian genomes and transcriptomes and recent evidence suggests that the proverbial ‘dark matter’ of the genome has major roles in cellular development, differentiation, and metabolism (10,12,13,24). Furthermore, accumulating reports of misregulated lncRNA expression across numerous cancer types suggest that aberrant lncRNA expression is a major contributor to tumorigenesis (12). NCRUPAR is a newly discovered long noncoding RNA molecule. Long Liu et al. demonstrated that NCRUPAR was significantly related to lymph node metastasis, distant metastasis, Duck’s stage, differentiation, and TNM stage in colorectal and gastric cancer (22,23).
In this study, we also discovered that the NCRUPAR expression level in five HCC cell lines, HCCLM3, HUH7, MHCC97H, SK-Hep1 and Hep3B, was significantly downregulated compared with a normal liver cell line QSG-7701 (Figure 1). Furthermore, it was downregulated in 73.7% of HCC tissues compared with the paired adjacent normal tissues (Figure 2). The Kaplan-Meier curve analysis showed that NCRUPAR expression were significantly related to overall survival of HCC patients (Figure 4). In summary, these findings imply that NCRUPAR may also acts as a tumor-suppressor lncRNA, which could affect cells phenotypically by promoting tumor-suppressor pathways, and when their function is compromised, cells are prone to develop cancer. In support of this notion, a few new studies have elucidated several examples of ‘tumor-suppressor lncRNAs’, such as LincRNA-p21, GAS5, and CCND1 (25,26).
Hepatocellular carcinoma is a complex, multi-factorial, multi-step disease. The overall survival of patients with HCC is grim because most patients are diagnosed late, when curative treatment is impossible. Clearly, there is a need for novel strategies for the early detection of HCC. Some molecular markers of HCC are important diagnostic and prognostic tools, which can help patient management (24,27,28). However, a clear advantage in the diagnostic use of ncRNA detection versus that of protein-coding RNAs is that in the former the RNA itself is the effector molecule, so its expression levels might be a better indicator of the intrinsic tumor characteristics. It is well known that the poor prognosis and high recurrence rate of HCC are due largely to the high incidence of intrahepatic and extrahepatic metastases (29). There is increasing evidence that lncRNAs play important roles in the invasion and metastasis of HCC (20,30,31). Our results showed that the NCRUPAR expression level was associated with cancer distal metastasis, portal vein tumor thrombus and histopathological grade (Table 1). These close correlations suggest that NCRUPAR is a potential target for gene therapy and could be used as a biomarker for predicting the prognosis of HCC. In addition, NCRUPAR is upstream from the PAR-1 gene and upregulates its expression during embryonic growth (18). However, Long Liu et al. demonstrated that NCRUPAR inhibits gastric and colorectal cancer progression by downregulating PAR-1 (19,20). Kaufmann et al. also demonstrated that the thrombin receptors PAR-1, PAR-3, and PAR-4 are expressed in HCC cell lines and that PAR-1 is involved in regulating hepatoma cell migration (20). To our knowledge, however, no authors have suggested how NCRUPAR regulates the PAR-1 gene in HCC and additional exploratory and validation research is needed to elucidate the functional role of NCRUPAR in HCC.
Our study had several limitations. Further understanding of the molecular mechanisms involving NCRUPAR in human HCC should facilitate the discovery of novel targeted agents and might also lead to the development of new approaches for effective therapy for human HCC. In order to explore the function of the NCRUPAR in vitro, lentivirus encoding the complete NCRUPAR open reading frame (LV-NCRUPAR) and a lentivirus vector encoding green fluorescent protein (LV-GFP), which was used as the control, were constructed, we detect the functional role of NCRUPAR in HCC cell lines, but the results is not well. The main mechanisms of lncRNA function, you know, such as HOTAIR, may serve as a scaffolding base for the coordination of epigenetic or histone-modifying complexes, including Polycomb repressive complexes and LSD1/CoREST and so on. When we constructed the lentivirus that encoding the complete lncRNAs open reading frame, the structures of the lncRNAs that has expressed by the artificially synthesized lentivirus may not be the original three-dimensional structure that exists in the cell and the post-transcriptional modification is also different. That is why most of researchers are only focus on the lncRNAs that highly expressed in the tumors, they can knock down or knock out the lncRNAs and not to overexpress the lncRNAs to research the function and mechanism of the lncRNAs. This can also explain why we cannot get the positive results between LV-NCRUPAR and LV-GFP in the HCC cell lines. Although many important questions remain unanswered, lncRNAs are shedding new light on our understanding of these cancer pathways; the potential roles of lncRNAs in biology and medicine could be tremendous; they might be useful as novel diagnostic and prognostic markers for various cancers. In addition, they might have therapeutic applications, although it will require many years of intensive research before they can be fully deciphered and applied. As more examples of regulation by lncRNA are uncovered, one might predict that the large transcripts will eventually rival small RNAs and proteins in their versatility as regulators of genetic information.
Conclusions
Our results indicated that NCRUPAR was downregulated in HCC and it could function as a potential biomarker in the diagnosis and estimation of the prognosis of HCC.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.03.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Human Research Ethics Committee of The First Affiliated Hospital, College of Medicine, Zhejiang University. Written informed consent was obtained from all subjects.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74-108. [Crossref] [PubMed]
- Yuen MF, Hou JL, Chutaputti A, et al. Hepatocellular carcinoma in the Asia pacific region. J Gastroenterol Hepatol 2009;24:346-53. [Crossref] [PubMed]
- Bruix J, Sherman MPractice Guidelines Committee, et al. Management of hepatocellular carcinoma. Hepatology 2005;42:1208-36. [Crossref] [PubMed]
- Ling TC, Kang JI, Bush DA, et al. Proton therapy for hepatocellular carcinoma. Chin J Cancer Res 2012;24:361-7. [Crossref] [PubMed]
- Llovet JM, Bruix J. Systematic review of randomized trials for unresectable hepatocellular carcinoma: Chemoembolization improves survival. Hepatology 2003;37:429-42. [Crossref] [PubMed]
- Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet 2003;362:1907-17. [Crossref] [PubMed]
- Shrimal A, Prasanth M, Kulkarni AV. Interventional radiological treatment of hepatocellular carcinoma: an update. Indian J Surg 2012;74:91-9. [Crossref] [PubMed]
- Tung-Ping Poon R, Fan ST, Wong J. Risk factors, prevention, and management of postoperative recurrence after resection of hepatocellular carcinoma. Ann Surg 2000;232:10-24. [Crossref] [PubMed]
- Qin LX, Tang ZY. The prognostic molecular markers in hepatocellular carcinoma. World J Gastroenterol 2002;8:385-92. [Crossref] [PubMed]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629-41. [Crossref] [PubMed]
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 2011;1:391-407. [Crossref] [PubMed]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol 2011;21:354-61. [Crossref] [PubMed]
- Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013;193:651-69. [Crossref] [PubMed]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43:904-14. [Crossref] [PubMed]
- Yang Z, Zhou L, Wu LM, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol 2011;18:1243-50. [Crossref] [PubMed]
- Niinuma T, Suzuki H, Nojima M, et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res 2012;72:1126-36. [Crossref] [PubMed]
- Wang F, Li X, Xie X, et al. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett 2008;582:1919-27. [Crossref] [PubMed]
- Braconi C, Kogure T, Valeri N, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011;30:4750-6. [Crossref] [PubMed]
- Yang F, Zhang L, Huo XS, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 2011;54:1679-89. [Crossref] [PubMed]
- Yuan SX, Yang F, Yang Y, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology 2012;56:2231-41. [Crossref] [PubMed]
- Madamanchi NR, Hu ZY, Li F, et al. A noncoding RNA regulates human protease-activated receptor-1 gene during embryogenesis. Biochim Biophys Acta 2002;1576:237-45. [Crossref] [PubMed]
- Liu L, Yan B, Yang Z, et al. ncRuPAR inhibits gastric cancer progression by down-regulating protease-activated receptor-1. Tumour Biol 2014;35:7821-9. [Crossref] [PubMed]
- Yan B, Gu W, Yang Z, et al. Downregulation of a long noncoding RNA-ncRuPAR contributes to tumor inhibition in colorectal cancer. Tumour Biol 2014;35:11329-35. [Crossref] [PubMed]
- Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol 2013;26:155-65. [Crossref] [PubMed]
- Coccia EM, Cicala C, Charlesworth A, et al. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol Cell Biol 1992;12:3514-21. [Crossref] [PubMed]
- Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010;142:409-19. [Crossref] [PubMed]
- Hoshida Y, Toffanin S, Lachenmayer A, et al. Molecular classification and novel targets in hepatocellular carcinoma: recent advancements. Semin Liver Dis 2010;30:35-51. [Crossref] [PubMed]
- Marrero JA, Lok AS. Newer markers for hepatocellular carcinoma. Gastroenterology 2004;127:S113-9. [Crossref] [PubMed]
- Budhu A, Forgues M, Ye QH, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell 2006;10:99-111. [Crossref] [PubMed]
- Yang F, Huo XS, Yuan SX, et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell 2013;49:1083-96. [Crossref] [PubMed]
- Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014;25:666-81. [Crossref] [PubMed]