
Next-generation sequencing–based clinical testing for lung cancer in Japan
Introduction
Recent insight into the molecular basis of lung cancer has led to changes in the treatment of this disease. The identification of driver genetic changes, such as those affecting the epidermal growth factor receptor (EGFR) (1-3) and anaplastic lymphoma kinase (ALK) genes (4,5), has already been successfully translated into clinical practice with the implementation of treatment with approved targeted agents (erlotinib, gefitinib, and afatinib for activating mutations of EGFR, and crizotinib and alectinib for rearrangements of ALK). The subsequent discovery of ROS1 and RET rearrangements as oncogenic drivers and potential therapeutic targets has shown that several chromosomal translocations and corresponding gene fusions can give rise to non–small cell lung cancer (NSCLC) (6-9). Given that mutations of EGFR, or KRAS and rearrangements involving ALK appear to be mutually exclusive, the need to test for many genetic alterations with a limited amount of collected tissue poses a problem for the “one companion diagnostic–one drug” paradigm of targeted therapy as applied to lung cancer and current clinical practice. The clinical implementation of genomic profiling for NSCLC with high-throughput and multiplex genotyping tests is thus urgently required in order to allow prioritization of appropriate therapies for individual patients.
The Lung Cancer Mutation Consortium (LCMC) evaluated the feasibility of simultaneously testing for 10 driver genetic alterations in patients with lung adenocarcinoma at 14 institutions in the United States (10). The study found that 64% of patients had one or more driver genetic alterations, and specific targeted therapy was administered to about half of these individuals. The median overall survival was longer for patients with a driver alteration who were treated with a driver-specific tyrosine kinase inhibitor than for those without an actionable genetic change or those with an actionable change who did not receive targeted therapy. These findings thus demonstrate the importance of simultaneous testing for multiple driver genetic alterations before administration of therapy in patients with advanced lung cancer.
Amplicon-based next-generation sequencing (NGS), as opposed to conventional Sanger-based sequencing, has been introduced to facilitate the performance of multiple genomic tests on the small amount of tissue available in a single formalin-fixed, paraffin-embedded (FFPE) tumor specimen obtained by transbronchial lung biopsy. This approach allows for the detection of rare as well as common cancer-related mutations that are clinically actionable with a short turnaround time and in multiple patients simultaneously, with a corresponding reduction in sequencing costs. In Japan, several institutions including the National Cancer Center and university hospitals have initiated NGS-based clinical testing and are able to provide access to investigational drugs or approved targeted agents matched to molecular alterations. In this review, we will briefly introduce the NGS-based clinical testing projects ongoing in Japan—in particular, as they apply to lung cancer—and we will discuss issues relating to the integration of NGS into clinical practice.
Genetic testing according to Japan Lung Cancer Society (JLCS) guidelines
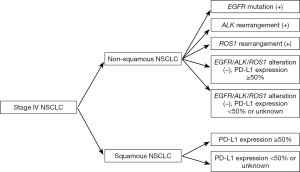
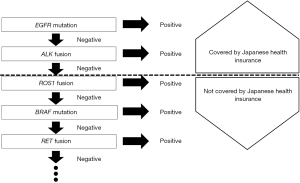
A revised version of clinical practice guidelines for the treatment of lung cancer in Japan was published by JLCS in 2016 (Figure 1), with the first decision step being based on histology (non–squamous cell carcinoma vs. squamous cell carcinoma) and, in the case of non–squamous cell carcinoma, the second decision step relating to the detection of EGFR mutations, ALK fusions, or ROS1 rearrangements. The cost of testing for EGFR mutations and ALK fusions is covered by the national health insurance system in Japan (Figure 2). As of March 2017, crizotinib has not been approved by the Pharmaceuticals and Medical Devices Agency (PMDA), a governmental organization in Japan, for the treatment of ROS1 rearrangement–positive lung cancer. However, it is listed in the current JLCS guidelines as an emerging targeted agent for patients whose lung tumors harbor such gene fusions, given that Pfizer filed an application in August 2016 seeking an additional indication of ROS1 fusion–positive advanced or recurrent NSCLC for this drug in Japan. For the subgroup of triple-negative patients (those negative for EGFR, ALK, and ROS1 changes), treatment decisions are made on the basis of the expression of programmed death-ligand 1 (PD-L1), given that pembrolizumab, a monoclonal antibody targeted to this protein, is approved for patients with NSCLC positive for PD-L1 expression at a threshold level of ≥50%. For patients with NSCLC tumors positive for the V600E mutation of BRAF, the combination of a BRAF inhibitor (dabrafenib) and an inhibitor of the downstream kinase MEK (trametinib) has been found to be highly effective as a therapy (11), and these drugs are currently under review by PMDA for the treatment of patients with unresectable or metastatic NSCLC positive for this BRAF mutation. However, BRAF testing is not currently listed in the revised JLCS guidelines.
SCRUM Japan
In February 2013, a large nationwide genomic screening program (Lung Cancer Genomic Screening Project for Individualized Medicine in Japan, or LC-SCRUM-Japan) was initiated with the aim of identifying patients with advanced non-squamous NSCLC negative for EGFR mutations but positive for rare driver genes and adapting molecularly targeted therapy accordingly. In November 2013, this program was expanded to also screen for driver mutations in fusion-negative tumors. In the screening protocol, genomic DNA extracted from biopsy or cytology specimens is analyzed with the Ion Torrent AmpliSeq Cancer Hotspot Panel version 2, which allows the simultaneous analysis of ~2,800 hotspot mutations in 50 cancer-related genes. As of May 2016, more than 200 institutions across the country were participating in LC-SCRUM-Japan. The program was transformed into a nationwide multicenter multiple-organ cancer genome screening network designated SCRUM-Japan. SCRUM-Japan now consists of the existing LC-SCRUM program and a gastrointestinal (GI) cancer screening network, GI-SCREEN. Patient specimens are collected from the ~200 participating institutions under local institutional review board approval and are now analyzed with a quality-assured Oncomine Cancer Research Panel (OCP, Thermo Fischer Scientific) at Clinical Laboratory Improvement Amendments (CLIA)–equivalent clinical laboratories. OCP detects amplifications and translocations as well as point mutations affecting ~140 cancer-related genes within 2 weeks. Clinical information and gene annotation for each patient are stored at the SCRUM-Japan data center in order to establish an interactive database, which will be open to collaborating researchers in academia and industry so as to enhance the research and development of new cancer diagnostics and treatments.
An example of the successful implementation of a novel trial model is provided by the LURET study (UMIN000010095), a phase II study of vandetanib in patients with RET-rearranged advanced NSCLC that is linked to SCRUM-Japan (12). Between February 2013 and March 2015, 1,536 patients with EGFR mutation–negative NSCLC were screened, of whom 34 individuals were found to be RET fusion positive (2%) and 19 were enrolled. Among 17 eligible patients included in the primary analysis, nine (53%) achieved an objective response, which met the primary end point of the study. Among the intention-to-treat population of all 19 patients treated with vandetanib, nine (47%) achieved an objective response. At the data cutoff, median progression-free survival was 4.7 months and no serious adverse events were noted, suggesting that vandetanib possesses antitumor activity and a manageable safety profile for patients with RET-rearranged advanced NSCLC and that RET rearrangement defines a new molecular subgroup of NSCLC amenable to targeted therapy.
Clinical sequencing at Kindai University
In July 2013, we launched NGS testing for lung cancer patients recruited from individuals set to undergo either biopsy for the purpose of diagnosis or curative surgery, or from others with already histologically proven lung cancer, at Kindai University Hospital (UMIN000014782). Invasive cancer cells occupying
Early results of this ongoing observational study have been published (13). Specimens from 110 patients with lung cancer were considered adequate for genomic testing between July 2013 and March 2015. Although most (64%) of these specimens were obtained by transbronchial biopsy and were small in size, the DNA and RNA samples extracted from the FFPE tumor tissue were successfully genotyped with amplification efficiencies of >90% (95% and 96% for DNA and RNA sequencing, respectively). The most common genetic alterations were TP53 mutations in 42 (38%) patients, followed by EGFR mutations in 25 (23%), STK11 mutations in 12 (11%), KRAS mutations in 10 (9%), MET mutations in 7 (6%), SMAD4 mutations in 7 (6%), and ERBB4 mutations in 4 (4%). Actionable genetic alterations were identified in 44 (40%) of the 110 study patients and included mutations in AKT1, BRAF, EGFR, KRAS, NRAS, PIK3CA, and STK11 as well as ALK, RET, and ROS1 fusions. Among the 37 patients with advanced or recurrent lung cancer harboring actionable alterations, 23 individuals (62%) received targeted therapy, including that with either clinically approved (n=16) or experimental (n=7) agents.
Patients with an actionable alteration who were treated with a targeted agent had a significantly longer overall survival compared with those without an actionable genetic change or those with an actionable change who did not receive targeted therapy. However, limitations of our study include its observational and nonrandomized design for comparison of overall survival between patients with tumors with an identified oncogenic driver according to whether or not they received a targeted therapy. Nevertheless, our findings indicate that such multiplex genomic testing—even with a single, small FFPE tumor specimen obtained by transbronchial lung biopsy—will assist physicians in matching patients found to harbor actionable mutations with available targeted treatments or clinical trials of new targeted agents.
OncoPrime Consortium in academia
In April 2015, Kyoto University Hospital launched a study of the clinical application of a CLIA-certifed comprehensive cancer gene panel (OncoPrime) in order to facilitate delivery of targeted therapies and thereby to improve outcomes for patients with rare cancers, primary unknown cancers, or cancers that failed standard chemotherapy (14). OncoPrime requires 100 ng of tumor DNA (obtained from 10 FFPE slides) and covers mutations in >200 cancer-related genes and rearrangements of 17 such genes. In addition to offering patients a matched targeted therapy, the goal of the study is to investigate the efficacy and clinical utility of comprehensive cancer genome analysis by NGS. The sequencing service for gene alterations in cancer tissue has since been expanded to include Okayama, Hokkaido, and Chiba University Hospitals.
MSK-IMPACT platform in academia
A clinical sequencing project named Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) was launched by Juntendo University Hospital in June 2016 and Yokohama City University Hospital in November 2016. This study has adopted a hybridization capture–based NGS assay for targeted deep sequencing of >400 cancer-related genes and 18 gene rearrangements, with the assay being performed in a CLIA-certified laboratory (15). MSK-IMPACT has obtained genetic information for >10,000 cancer patients. The median processing time from informed consent to reporting the genotyping results to the treating physician is ~1.5 months.
Issues relating to the clinical application of NGS panels in Japan
The accumulating operational experience with the application of NGS-based genetic testing in Japan has raised several concerns. Most clinical NGS tests are currently performed as Laboratory Developed Tests (LDTs), as there is no PMDA-approved NGS platform for detection of somatic mutations. Given that the development of a custom NGS test requires a substantial investment in operational and bioinformatics infrastructure and that such investment may not be feasible for many clinical laboratories, most Japanese groups have opted to validate ready-made vendor solutions from outside Japan, such as the Ion Torrent Cancer Panel (Life Technologies) and TruSeq Amplicon Cancer Panel (Illumina). Moreover, there is no government support to develop a domestic NGS panel or to promote innovation by Japanese industry. The fact that some samples are shipped overseas for NGS testing has raised suspicions that the genotyping data may be used for other purposes such as the development of single nucleotide polymorphism (SNP) or copy number variation (CNV) databases for the Japanese population. There is an urgent need to develop novel agents for cancer therapy and to guide patients to clinical trials evaluating drugs matched to identified molecular alterations in Japan. Given that recent targeted drugs have been approved together with corresponding clinically validated companion diagnostic tests, somatic variants identified by NGS used for research purposes cannot serve to directly guide patient care. An educational curriculum has been established for inherited disorders of the nervous system and of metabolism. Hereditary breast and ovarian cancer (HBOC) is a cancer susceptibility syndrome involving breast, ovarian, or prostate cancers caused by germline mutations of BRCA1 or BRCA2, and a Japanese HBOC consortium was established for the purpose of patient registration and the provision of essential genetic information in clinical practice. On the other hand, there is no established medical curriculum for interpretation and annotation of NGS data obtained from cancer patients in Japan.
Conclusions
Clinical application of amplicon-based NGS to the performance of multiple genomic tests with a single, small FFPE tissue specimen provides a large amount of information regarding the genetic alterations of the corresponding tumor. Such multiplex genomic testing helps physicians to match patients found to harbor actionable alterations with available targeted treatments or clinical trials of new targeted agents. Further development of this field will require real-time knowledge of genomic alterations that can be used in clinical decision making.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.04.12). MT serves as an unpaid editorial board member of Translational Cancer Research from Nov 2016 to Dec 2018. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306-11. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Takeda M, Okamoto I, Sakai K, et al. Clinical outcome for EML4-ALK-positive patients with advanced non-small-cell lung cancer treated with first-line platinum-based chemotherapy. Ann Oncol 2012;23:2931-6. [Crossref] [PubMed]
- Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med 2012;18:375-7. [Crossref] [PubMed]
- Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012;18:378-81. [Crossref] [PubMed]
- Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012;18:382-4. [Crossref] [PubMed]
- Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012;30:863-70. [Crossref] [PubMed]
- Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998-2006. [Crossref] [PubMed]
- Planchard D, Kim TM, Mazieres J, et al. Dabrafenib in patients with BRAF(V600E)-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol 2016;17:642-50. [Crossref] [PubMed]
- Yoh K, Seto T, Satouchi M, et al. Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): an open-label, multicentre phase 2 trial. Lancet Respir Med 2017;5:42-50. [Crossref] [PubMed]
- Takeda M, Sakai K, Terashima M, et al. Clinical application of amplicon-based next-generation sequencing to therapeutic decision making in lung cancer. Ann Oncol 2015;26:2477-82. [PubMed]
- Kou T, Kanai M, Matsumoto S, et al. The possibility of clinical sequencing in the management of cancer. Jpn J Clin Oncol 2016;46:399-406. [Crossref] [PubMed]
- Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17:251-64. [Crossref] [PubMed]


