
Epigenetic mutations and cancer therapy Effectiven(EZH2)
Bladder cancer (BC) as epigenome disease
Cancer arises as a consequence of accumulating genomic alterations, which affect primarily oncogenes and tumor suppressor genes (TSGs). This condition creates a particular landmark that can be exploited in therapy. Not surprisingly, many pharma companies launched programs aimed to inhibit the gain of function activities caused by oncogene mutation and/or amplification, and many of these inhibitors are currently in the clinics. The situation is not so simple in the case of TSGs, where a common loss of function precludes, in most cases, their actual use as therapeutic targets.
BC is a clinical challenge due to its incidence, prevalence and mortality rates (1). Moreover it also represents an economic problem as, due to its high rate of recurrence, a regular surveillance by cystoscopy and urine cytology is required (2). BCs are also characterized by the frequent alterations in genes governing chromatin organization and histone modifications, as shown by whole exome sequencing studies, leading to particular changes in the epigenome reflected in altered expression of multiple genes (3). Chromatin regulatory elements, which are epigenetic molecules that regulate flexible processes based on little post-transcriptional modifications such as acetylation, methylation or ubiquitination to express or repress genes, appeared therefore highly altered in BC and, within these alterations, at least 89% are histone remodelers and 64% nucleosome positioning genes (4).
The role of epigenetics is surprisingly wide and involves not only chromatin remodelers, but also changes in DNA methylation, expression of miRNAs, lncRNAs, etc. Therefore, epigenetic factors are becoming attractive targets to develop new treatments for patients. Regarding chromatin remodelers in BCs, it is worth considering the two main pathological entities of these diseases: the non-muscle invasive BC (NMIBC), and muscle invasive BC (MIBC). While this pathological classification is now under question due to the discovery of intrinsic subtypes (5), through whole transcriptome studies, similar to other solid tumors, it also has huge relevance as it currently defines the possible therapeutic options. The NMIBC are treated by transurethral resections, in some cases followed by intravesical therapy, whereas the MIBC are regularly treated by cystectomy followed by chemotherapy. Although NMIBCs have a more favorable prognosis, they frequently recur and acquire MIBC features. Therefore, it is necessary to identify and characterize precise biomarkers of recurrence and progression for early diagnosis and follow-up, which may become possible therapeutic targets and be a non-surgical treatment option, improving patient survival and prevention of tumor recurrence. The aberrant epigenetic landscape is a hallmark of human cancer (6) and characterizes BC as an epigenome disease. The identified epigenetic alterations suggest new possibilities for the treatment of different bladder tumors, making them good candidates for epigenetic therapy.
Therapy for synthetic lethality in suppressor genes
Synthetic lethality occurs when the expression of two genes (or their mutated counterparts) promotes cell death. Usually these two genes are not co-expressed, are mutually exclusive, and conforming a lethal interaction. These lethal interactions are based either on genetic mutations or the introduction of molecules with a known cytotoxic effect in cancer cells or patients. The acquisition of mutations confers tumor cell advantages in proliferation, survival and even drug resistance, attributing them a pattern of differential gene regulation to normal cells. This hallmark can convert tumor cells into therapeutic targets, identifying those alterations that can be targeted to induce specifically synthetic lethality (7). Therefore, the use of synthetic lethality strategies would bring us closer to more targeted treatments for different patients (5). Nowadays, progress is being made in synthetic lethality of genes such as RB1, TP53, BRCA1, RAS and C-Myc (8).
There are different experimental methodologies to identify the inactivation of genes that can show a lethal phenotype under a given genotype. Currently, in some cancers with TSGs mutants, large libraries of small hairpin RNA (shRNA) (9), small interfering RNA (siRNA) (10), and RNA guides for gene editing using CRISPR/Cas9 or TALENs (11) are commonly used. The main difference between these techniques is the inactivation efficiency: while CRISPR engineering allows for complete silencing of genes, the use of shRNA or siRNA induces a downregulation of the genes that is temporary and with greater variability between recognizable sequences. These small RNAs can recognize other target sites, known as off-targets, which distort the interpretation of data, creating false positive genes for synthetic lethality (9). Regarding CRISPR/Cas9 approaches, they may lead to side off effects due to the guides selected. However, CRISPR/Cas9 can be so selective and efficient that the inactivation of an essential gene is lethal per se, and therefore does not allow to see the phenotype that can be observed with the use of shRNA. In addition, most of the drugs in development are competitive inhibitors, so that partial inhibition promoted by shRNA allows for the reproduction of the pharmacological effect.
In BC, substantial evidence that the epigenome shows profound alterations is reflected in a frequent silencing in some well-known TSGs such as histone demethylase KDM6A, which is mutated in 24% of BC cases, or ARID1A, which is part of the SWI/SNF complex of nucleosomes and altered in 25% of BC cases (6). Mutations in these tumor suppressors, often deep deletions or putative truncating mutation (4), may help identify patients susceptible to synthetic lethality, one of the most promising recent approaches in epigenetic therapy. In different tumor subtypes in which there is already a silenced gene, an interesting possibility could be to take advantage of this silencing to perform the inhibition of an antagonist of KDM6A and ARID1A tumor suppressors, like EZH2, to promote synthetic lethality. EZH2, a histone methyltransferase that regulates gene silencing through the H3K27me3 mark, is part of the polycomb repressive complex 2 (PRC2) together with other factors such as EED or SUZ12. Upregulated EZH2 can inactivate the transcription of many other suppressor genes, thus having an oncogenic functionality. Interestingly, EZH2 is involved in the processes of recurrence and progression in NMIBC (12). Furthermore, different studies have found that the suppressor genes KDM6A and ARID1A antagonistically regulate the expression of genes involved in cell proliferation and survival, such as PIK3IP1, which negatively regulates the PI3K-AKT pathway, and IGFBP3, which is involved in anti-proliferative signals, appears frequently silenced in BC, and is activated by inhibiting EZH2 (10,11).
Therefore, synthetic lethality emerges as a novel approach in targeted therapy for BC, as well as many other cancers, as it reduces the secondary effects of chemotherapies, tackles drug resistance, and improves knowledge of signaling pathways to define more accurately the different tumor subtypes. Moreover, synthetic lethality provides an additional way for the individualized treatment of patients (9).
The complexity of EZH2 as treatment target: mutations, different roles and acquired resistance
Among the most altered chromatin remodelers altered in BC, inactivating mutations in the histone H3 lysine 27 (H3K27) demethylase KDM6A (also known as UTX) were most common and enriched in NMIBCs (32–43%) (1,3), whereas inactivating mutations in the SET family histone H3 lysine 4 (H3K4) methyltransferase MLL2 were more common in MIBCs (19%), and mutations in KDM6A and MLL2 were mutually exclusive (1). This fact is not well understood at present and would require in depth chromatin immunoprecipitation/sequencing (ChIP-seq) future studies.
KDM6A gene is a histone H3 trimethyl (H3K27me3) demethylase located on chromosome Xp11.2. This gene plays specific functions creating a transcription-permissive chromatin structure through its demethylase activity, and displays multiple alterations in a wide range of human tumors, in most cases leading to loss of function situation (Figure 1A,B). This has led to the assumption that KDM6A represents a bona fide TSG in the BC context. The KDM6A function opposes to that of EZH2, which is the catalytic subunit of PRC2, frequently overexpressed in multiple malignancies (prostate, breast, bladder, endometrial, melanoma, etc.) showing a positive correlation with high grades and worse prognosis (1,12,13). This frequent overexpression has also promoted the increased interest in developing EZH2 inhibitors still presently in clinical trials (1,13,14).
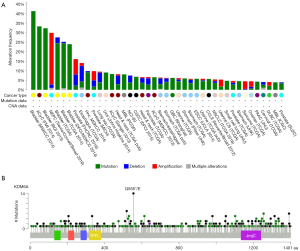
In addition, the functional antagonism between KDM6A and EZH2 has been recently exploited to find possible new avenues in BC therapy. In a recent elegant study, Ler et al. have confirmed that KDM6A is frequently lost in MIBC and NMIBC accounting for the enrichment in PRC2-regulated signaling (11). Remarkably through various experimental approaches, they also showed that loss of KDM6A confers specific vulnerabilities to EZH2 inhibition. In particular, EZH2 inhibition delays tumor onset and induces tumor regression of KDM6A-null cells and patient-derived xenografts models. This work represents an excellent example of how TSG loss vulnerabilities can be exploited in the context of cancer therapy. Of note, similar vulnerability to EZH2 inhibitors has been previously reported for ovarian tumors bearing mutations in ARID1A gene (10). ARID1A encodes a component of the SWI/SNF chromatin-remodeling complex, and also shows a high mutation rates in multiple cancer types, thus playing various TSG functions. Similarly, loss of ARID1B in ARID1A-deficient backgrounds destabilizes SWI/SNF and impairs proliferation in cancer cells (15). The SWI/SNF complex is composed by the combinatorial assembly of around 15 subunits and contributes to DNA repair and to transcriptional regulation in a lineage-specific manner. Since genetic approaches in various organisms, including fly, showed opposite roles for SWI/SNF and Polycomb mutations, the possible susceptibility of tumors bearing ARID1A, PBRM1 and SMARCA4 alterations to EZH2 inhibition has also been tested (7). Importantly SWI/SNF-mutant cancer cells are primarily dependent on a non-catalytic role of EZH2, being only partially dependent on EZH2 histone methyltransferase activity (7).
This is also relevant in the context of BC. Indeed SWI/SNF-complex subunits are also frequently altered in BC patients (Figure 2) (4) with various mutual exclusivity/co-occurrence situations. According to this, EZH2 is an attractive target for BC management. However, further elucidation and research on this aspect is extremely needed. On one hand, among the various EZH2 inhibitors under study some are catalytic inhibitors with pleiotropic actions due to their action as SAM-hydrolase or SAM-competitive inhibiting compounds (16), whereas others may affect the stability of PRC2 complexes. Accordingly, the complete determination of the dependence on EZH2 catalytic activity in the various mutated genes is required. On the other hand, the specificity of these inhibitors over EZH2 or EZH1 is strictly required. Indeed, these two proteins may play overlapping but also different functions in normal organism development and in cancer (17). In addition, it is worth to mention that several studies have demonstrated that specific EZH2 inhibitors seem to be particularly effective in impairing cell growth in a mutant EZH2 background, showing a low effectiveness in those cases with wild type EZH2 (16,18). It is also important to note here that effects of EZH2 catalytic inhibitors are really time-dependent, requiring a long-term treatment due to the slow kinetics of H3K27me3 turnover, thus highlighting the requirement for deep pharmacokinetic and pharmacodynamics in addition to toxicity studies.

Another aspect to consider is the existence of EZH2 mutations. While these have been considered rare in BC, they are frequent in other tumor types, such as various hematological tumors. Whereas in general the mutations lead to a gain-of-function of its enzymatic activity (19), in some other the mutations account for a loss of function, thus suggesting a possible TSG functions of EZH2 (20). In fact, inactivating and loss-of-function mutations in EZH2 have been reported in myelodysplastic syndrome (MDS), myeloproliferative neoplasms (MPN), MDS-MPN overlap disorders, and T-cell acute lymphoblastic leukemia (T-ALL) (1). Interestingly, in T-ALL for instance, Ntziachristos et al. have demonstrated an interplay between an oncogenic role of NOTCH1 and tumor suppressor role for PRC2, opening a new therapeutic possibility by combining inhibitors of H3K27 demethylases and targeted anti-NOTCH1 therapies (21). These apparent discrepancies could be ascribed to different functions of EZH2 in several tissue types, implying that EZH2 role is not simply promote either stemness or differentiation per se, and leading to the idea that targeting its expression can have consequences highly cell type specific. Also in this regard, EZH2 gain-of-function, produced either by mutations or through inactivating mutations affecting others chromatin regulators antagonizing EZH2 activity, such as KDM6A mutations, can account for methylation of non-histone substrates, presenting a PRC2-independent function (22), such as those transcriptional activations mediated by AR in prostate cancer, by NF-kB or NOTCH1 in breast cancer, or by ER and WNT signaling transcription factors (1). The fact that loss of function in PRC2 genes or its substrate H3K27 is associated with oncogenesis highlights the need of being really cautious related to the use of EZH2 inhibitors in the clinic, since the possible long-term therapies needed would result in increased incidence of undesired secondary effects, including hematological malignancy development.
Considering the need of the long-term cancer treatment, the possible acquired resistance becomes a main problem. The resistance can be acquired by amplification or secondary mutations of the drug targets, or through activation of bypass signaling pathways (14). Also other epigenetic factors, such as EHMT2, have been shown to be able of compensating the loss of EZH2. Accordingly, careful studies aimed to understand the resistance mechanisms are needed. In this sense, using EZH2-mutated lymphoma cells, Gibaja et al. developed models of resistance to EZH2 inhibitor EI1 by a prolonged exposure to the drug (14). Their results also supported a cooperation model between EZH2 WNT and Y641N mutants, highlighting the fact that only targeting EZH2 WT treatment could not be effective. Kim et al. also reported two novel secondary EZH2 mutations after a long inhibitory exposition in a cell line model, able of conferring resistance (1). These results implicate that new treatments should target both possibilities, EZH2 WNT or mutants, and more ideally also to EZH1, since it might also contribute to resistance. Finally, loss of PRC2 subunits has been reported to amplify Ras-driven transcription in different tumors, showing also a high correlation with resistance to EZH2 inhibition (1). Therefore, all these results point towards the essential development of new drugs or combined therapies, including different EZH2 inhibitors, in order to prevent, or bypass, the possible resistance mechanisms and achieve a more long-term effectiveness.
In conclusion, although the ongoing research provides new therapeutic options for the management of BC cancer patients based on specific mutations, and targeting epigenetic remodelers, a better understanding of their mechanistic roles is strictly required.
Acknowledgments
Funding: FEDER cofounded MINECO grant (SAF2015-66015-R), Comunidad Autónoma de Madrid Oncocycle Program grant (S2010/BMD-2470), MSyC grant ISCIII-RETIC (RD12/0036/0009, PIE 15/00076, CB/16/00228) to JM Paramio.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Peng Zhang (Department of Urology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.05.27). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kim J, Akbani R, Creighton CJ, et al. Invasive Bladder Cancer: Genomic Insights and Therapeutic Promise. Clin Cancer Res 2015;21:4514-24. [Crossref] [PubMed]
- Burger M, Oosterlinck W, Konety B, et al. ICUD-EAU International Consultation on Bladder Cancer 2012: Non–Muscle-Invasive Urothelial Carcinoma of the Bladder. Eur Urol 2013;63:36-44. [Crossref] [PubMed]
- Gui Y, Guo G, Huang Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet 2011;43:875-8. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315-22. [Crossref] [PubMed]
- Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer 2015;15:25-41. [Crossref] [PubMed]
- Han H, Wolff EM, Liang G. Epigenetic Alterations in Bladder Cancer and Their Potential Clinical Implications. Adv Urol 2012;2012:1-11.
- Kim KH, Kim W, Howard TP, et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med 2015;21:1491-6. [Crossref] [PubMed]
- Beijersbergen RL, Wessels LF, Bernards R. Synthetic Lethality in Cancer Therapeutics. Annu Rev Cancer Biol 2017;1:141-61. [Crossref]
- Leung AW, de Silva T, Bally MB, et al. Synthetic lethality in lung cancer and translation to clinical therapies. Mol Cancer 2016;15:61. [Crossref] [PubMed]
- Bitler BG, Aird KM, Garipov A, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med 2015;21:231-8. [PubMed]
- Ler LD, Ghosh S, Chai X, et al. Loss of tumor suppressor KDM6A amplifies PRC2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of EZH2. Sci Transl Med 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Santos M, Martínez-Fernández M, Dueñas M, et al. In vivo disruption of an Rb-E2F-Ezh2 signaling loop causes bladder cancer. Cancer Res 2014;74:6565-77. [Crossref] [PubMed]
- Comet I, Riising EM, Leblanc B, et al. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer 2016;16:803-10. [Crossref] [PubMed]
- Gibaja V, Shen F, Harari J, et al. Development of secondary mutations in wild-type and mutant EZH2 alleles cooperates to confer resistance to EZH2 inhibitors. Oncogene 2016;35:558-66. [Crossref] [PubMed]
- Helming KC, Wang X, Wilson BG, et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat Med 2014;20:251-4. [Crossref] [PubMed]
- Morera L, Lübbert M, Jung M. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin Epigenetics 2016;8:57. [Crossref] [PubMed]
- Margueron R, Li G, Sarma K, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 2008;32:503-18. [Crossref] [PubMed]
- Curry E, Green I, Chapman-Rothe N, et al. Dual EZH2 and EHMT2 histone methyltransferase inhibition increases biological efficacy in breast cancer cells. Clin Epigenetics 2015;7:84. [Crossref] [PubMed]
- Souroullas GP, Jeck WR, Parker JS, et al. An oncogenic Ezh2 mutation induces tumors through global redistribution of histone 3 lysine 27 trimethylation. Nat Med 2016;22:632-40. [Crossref] [PubMed]
- Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med 2016;22:128-34. [Crossref] [PubMed]
- Ntziachristos P, Tsirigos A, Van Vlierberghe P, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med 2012;18:298-301. [Crossref] [PubMed]
- Martínez-Fernández M, Rubio C, Segovia C, et al. EZH2 in Bladder Cancer, a Promising Therapeutic Target. Int J Mol Sci 2015;16:27107-32. [Crossref] [PubMed]


