Unsupervised clustering reveals new prostate cancer subtypes
Introduction
Despite the fact that the advent and innovation of the screening technology has made prostate cancer easier to be diagnosed, with an ever rising incidence, it has been the second cause of cancer-related death among American males (1-3). More than 95 percent of prostate cancer presents as adenocarcinoma. Thus, it is of great importance to explore the developing and prognostic stratification of prostate cancer, especially the molecular classification.
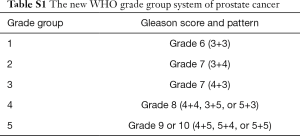
Based upon architectural features of prostate cancer cells, the Gleason score is an efficient way to evaluate the malignancy of prostate cancer. Traditionally, the Gleason score is calculated by adding together the numerical values of the two most prevalent differentiation patterns (a primary grade and a secondary grade). However, a revised grading system (Table S1) has been adopted in 2016 World Health Organization classification of genitourinary tumors (4,5). An increasing risk of biochemical recurrence (BCR) with elevating grade has been observed (6). Besides the clinical indexes, some protein-coding genes such as c-myc, Bcl-2 and p53 have also been reported to be related to the prognosis of prostate cancer over the last decades (7,8).
The Cancer Genome Atlas (TCGA) is a large project composed of multiple components and equipped with various custom applications to handle large volumes of research data (9). It contains genomic characterization data, high level sequencing data and corresponding clinical data of all common tumors and several rare tumors. The integrative analyses made by the TCGA research network in some tumors like glioma and ovarian cancer provided a new insight into their diagnoses and treatment strategies (10,11). A global analysis of prostate adenocarcinoma has been published by the TCGA research network, revealing molecular heterogeneity and potentially actionable molecular defects among primary prostate cancers (12).
As a subfield of computer science, machine learning plays an important role in bioinformatics for its incomparable advantage in handling big data. Up to now, it has made great contribution in the discovery of many valuable results (13,14). In the current study, we applied the previous methods to the TCGA database analysis, and found out some interesting results, which could be validated stably in three other datasets regardless of platforms.
Methods
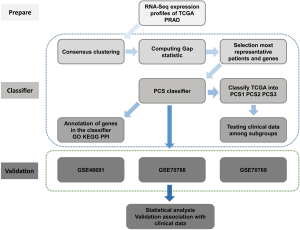
We summarized our workflow in the Figure 1.
Data preparing
We used four different series of prostate cancer data with 1258 unique patients in this study (Table S2). TCGA RNAseqV2 dataset consisted of 497 prostate adenocarcinoma samples as a training dataset. The validation datasets comprised three prostate cancer series from GEO (Gene Expression Omnibus): Erho’s series (GSE46691) with 545 patients (15), Lamb’s set (GSE70768) with 125 patients (16) and Ross’s set (GSE70769) with 91 patients (16) respectively.
The training dataset (TCGA RNAseqV2 and Clinical data)
Prostate adenocarcinoma level-3 data were obtained from TCGA consortium (https://tcga-data.nci.nih.gov/tcga/). This dataset included 497 individuals with prostate adenocarcinoma. The RNA sequences of each sample were profiled based on Illumina HiSeq 2000 RNA Sequencing Version 2 analysis (https://wiki.nci.nih.gov/display/TCGA/RNASeq+Version+2). Tumor samples from TCGA were not only in different institutions and distinct times, but also processed in batches rather than at the same time, which would lead to misleading analysis resulting from systematic noise such as batch effects and trend effects. TCGA BatchEffects website (http://bioinformatics.mdanderson.org/tcgambatch/) was used to assess and correct for batch effects in prostate adenocarcinoma data. We downloaded the batch effects corrected data processed by Empirical Bayes online. Besides, clinical data was downloaded through R package of TCGA-Assembler (17).
The validation dataset (GEO series)
The expression profiles as well as clinical data of GSE46691, GSE70768 and GSE70769 were downloaded through R package of GEOquery. For each dataset, the expression profiles were annotated from probesets to genes and median centred across all samples. We filled target gene expression with zero when missing value occurred. For clinical data, we concentrated on Gleason score, T staging and prognostic information.
Generation of the prostate cancer subtype (PCS) classifier and identification of subtypes
We built up a PCS classifier to identify three subtypes based on RNA-Seq expression profiles of TCGA with algorithm of Hierarchical clustering and nearest shrunken centroids.
Consensus clustering
Consensus clustering is one way of assessing the clustering stability. With the R package of “ConsensusClusterPlus” (18), we carried out hierarchical clustering with agglomerative average linkage and classified these patients into 3–12 clusters via the 10426 most variable genes. The most variable genes were defined according to the criterion of median absolute deviation >0.5. We had median centred all expression arrays before all the computations and set 1,000 iterations, 0.98 subsampling ratio for consensus clustering (19).
Computing gap statistic
Gap statistic is a standard method to determine the optimal number of clusters in a dataset via comparing the change of the observed and expected within-cluster dispersion (20). To identify the ideal number of clusters, gap statistic was computed from k=1 to 6 for selected top variable genes by the R package of “cluster” (21).
Selection of patients and genes for machine learning
For patient samples, we computed Silhouette width to recognize the most representative patients in each cluster (22). Patients with a positive silhouette width were selected for the following classifier. Similarly, two filtering steps were applied to select the most representative and predictive genes. Firstly, SAM (Significance Analysis of Microarrays) was used to identify significantly differentially expressed genes (FDR <0.01, False Discovery Rate) between one subtype and others with R packages of “siggenes” (23). Secondly, AUC (area under receiver operating characteristic curve curve) values were calculated to estimate one gene's predictive ability to divide one subtype from others with R packages of “ROCR” (24). Only patients with FDR <0.01 and AUC >0.9 were kept to build the PCS classifier.
Building the PAM classifier and identifying subtypes
With the filtered patients and selected genes, a robust classier was established by the R packages of “PAMR” (prediction analysis for microarrays R package) based on the algorithm of nearest shrunken centroids (25). We set up a 10-fold cross-validation for 1,000 iterations to choose the optimal threshold of centroid shrinkage. Finally, we selected the classifier providing error rate <2% with the least number of genes. As a result, with the built PAM classifier, we classified all the TCGA prostate adenocarcinoma patients into three subgroups for the subsequent analyses. Firstly, we fit the clinical data to the three subtypes of prostate adenocarcinoma to check if there would be a difference in Gleason score, T staging, or prognosis. Secondly, in order to explore the molecular heterogeneity, we selected some popular biomarkers or mutations to check their expression variation in the different subtypes.
Annotation of genes in the classifier
To understand the biological significance of PCS genes, we performed annotation for genes by the following methods.
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
Gene ontology annotation associated with biological pathway and KEGG pathway enrichment analysis of the classifiers was achieved using the Database for Annotation Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) online tool. A P value <0.001 and gene counts >2 were set as threshold for biological pathway analysis, and P value <0.05 and gene counts >2 for KEGG pathway analysis.
Protein-protein interaction (PPI) network construction
We mapped the classifiers into the Search Tool for the Retrieval of interaction Genes database (String, http://string.embl.de/) and constructed a PPI network, which provided the information to predict the protein interactions. The minimum required interaction score was set to 0.400. We analyzed the PPI network with Cytoscape software (http://www.cytoscape.org/).
Validation on three GEO dataset
GSE70769, GSE70768 and GSE46691 from GEO dataset were selected as validation sets. Based on the PCS classifier generated in section 2.2.4, we classified each validation dataset and analyzed the relation between the given subgroups and clinical data. Some clinical information of all series was not identical. For example, the GSE70768 gave the primary and secondary Gleason score, while the GSE46691 just gave total Gleason score. Concerning the issues above, we utilized the majority of clinical data and presented the details in the results part.
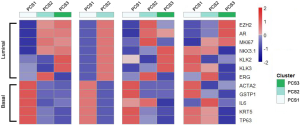
Expression of the luminal and basal markers in the PCS subtypes.
To determine whether the PCS subtypes correspond to luminal or basal tumors, we analyzed the mean expression of genes known to be markers of luminal (EZH2, AR, MKI67, NKX3-1, KLK2/3 and ERG) or basal (ACTA2, GSTP1, IL6, KRT5 and TP63) prostate cancers among PCS subtypes (26). We also performed the gene set enrichment analysis (GSEA) to provide some biological insights of the PCS clusters (27).
Statistical analysis
Sample clustering and classifying were performed with the corresponding R packages mentioned above under R software (version of 3.3). Clinical data were treated as discrete variable (time to event) and categorical variable (Gleason score, pathology T stage (pT stage), metastasis, and prognostic end-point). We utilized Chi-square test and fisher’s exact test when detecting the relation between categorical variables. Kaplan-Meier curves were used to describe the time-event data and log-rank method was used to test the differences. It was regarded as significant statistically when P value was <0.05. We applied the statistical packages SPSS v20 (IBM) to manage the clinical data. R software and Microsoft PowerPoint (v2016) were used for visualizing the results.
Results
Generation of the PCS classifier and subtypes identification
A total of 10,426 genes with most variability across samples were retained and median centred. Next, we performed hierarchical clustering with agglomerative average linkage to cluster these samples. We employed consensus clustering to assess the clustering stability. A significant increase in clustering stability was observed from k=2 to 3, but not for k>3 (Figure 2A). To further confirm it, we computed Gap statistic for k=1 to 5. A peak was mainly at k=3 or 4, indicating that three or four subtypes were ideal to explain the inner construction of dataset. To simplify the explanation for the result, we chose three subtypes for our following analysis (Figure 2A). We computed Silhouette width to identify the most representative samples within each cluster. To build the PCS classifier, we retained samples with positive silhouette width (n=412) (Figure 2A). We also applied two filtering methods (SAM and AUC) to select the most representative and predictive genes. The retained 256 genes were trained by PAM to build a classifier. To select the optimal threshold for centroid shrinkage, we performed 10-fold cross-validation and selected the one yielding a good performance (error rate <2%) with the least number of genes. Using this strategy, we built a classifier of 183 unique genes (Figure 2B) (Table S3—The gene list of PCS classifier). These genes were annotated with biologic process, biologic pathway, protein-protein interaction databases. Then we used it to classify all the prostate adenocarcinoma samples. 250 samples were classified for the first subtype (PCS1, prostate cancer cluster 1), 153 were PCS2, and the rest were PCS3 (Figure 2C). Some candidate genes were found to be validated in this dataset. They were KLF5, BCL2 and etc., and Figure 2B gave the details.
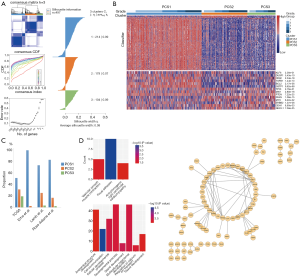
Function annotation and PPI analysis of our classifier
As partially displayed in Figure 2D, the classifier was mainly annotated with 48 biologic process and biologic pathway terms, most of which were concerning cell communication, development process, morphogenesis and ion transport. KEGG pathway analysis revealed that these classifiers mostly enriched in focal adhesion.
The PPI network included 86 nodes and 112 edges. Some genes validated by other research were in the centre of the network, such as SNAI2, FGFR2, SPEG, CAV1 and so on, which might serve as intermediate regulation. Other genes validated by former research included FHL2, CD40, ANPEP, GSTP1, FLNA (28-33), which were not so highly interconnected with other genes.
Analysis with clinical data
After statistical test with the TCGA clinical data, we found PCS2 with an obviously higher Gleason score when comparing to PCS1 (Figure 3A). PCS3 had the least number of patients, and seemed to be the buffer zone for PCS1 and PCS2. About 1/3 PCS1 patients were in the WHO grade group 2 (Gleason score at 7=3+4). As for the PCS2, more than 1/3 patients were in the group 5 (Gleason score at 10).
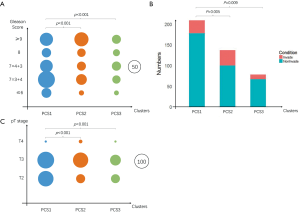
The pT stage and lymph node invasion had the same tendency (Figure 3B,C). PCS2 had higher levels of pT stage and N stage comparing to PCS1. However, the recurrent times had no significant difference among these subtypes, just like what the TCGA research network reported (12). The insufficient follow-up time (median follow-up time was 15.1 months) and low follow-up rate (less than 66%) might account for that.
Validation in three other datasets
The interesting results of above had been validated only in TCGA dataset. To push them to universal, we found three more datasets for further validation. Some information about these series was summarized in Table S2.
GSE46691
We applied the classifier to this dataset. Only two subtypes (PCS1 and PCS2) were found, but none for PCS3. Most of them were PCS1(532/545 vs. 13/545) (Figure 2C).
Different from TCGA dataset, this series provided too little information for us to use the WHO grading system. Thus, we separated all the patients to two levels with a cutoff of 8 (≤8 and >8). After comparison, Gleason scores and metastasis status had extremely similar patterns in these two different subtypes. The different results between PCS1 and PCS2 were shown in Figure 4A.
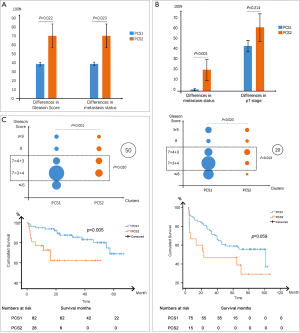
GSE70769
With the similar classification, there were only two patients in PCS3. Thus, we only analyzed PCS1 [74] and PCS2 [15] for the following items. The significant difference was observed between distinct levels of Gleason grading system (P=0.023) (Figure 4B). Unlike TCGA dataset, pT stages were not significantly different between PCS1 and PCS2 (Figure 4B). As for metastasis status, positive results could be observed (P=0.001), despite the paucity of metastatic cases (Figure 4B). Unfortunately, we observed no positive results concerning the recurrences between PCS1 and PCS2 in the survival analysis (P=0.059) (Figure 4B). However, the tendency of survival curve and critical P value implied a possibility of difference.
GSE70768
After classification, 90 cases were tagged as PCS1, while 31 were in PCS2 and four were in PCS3. Following GSE70769, we ignored PCS3 for further analysis. Besides the Gleason scores, the recurrence time was also significantly different between PCS1 and PCS2 in the survival analysis (P=0.005) (Figure 4C).
Expression of the luminal and basal markers in the PCS subtypes
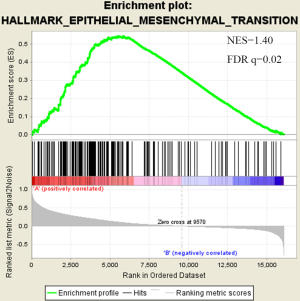
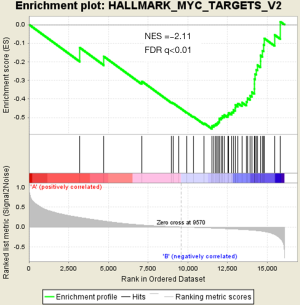
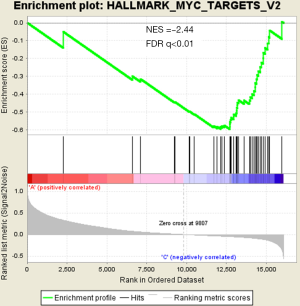
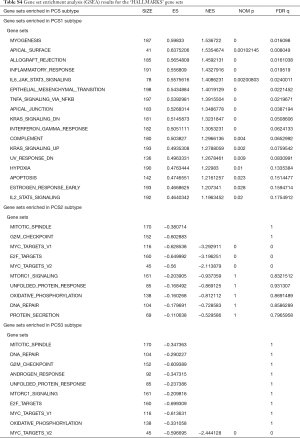
There were an association between luminal genes and “PCS2 and PCS3”, and basal genes and PCS1 (P<0.05). We also verified this observation in other 3 data sets. It provides evidences that PCS1 tumors correspond to basal subtypes, PCS2 and PCS3 reflect luminal subtypes. Table S4 shows the gene sets enriched in each of PCS subtypes by GSEA. There are 17 gene sets significant enriched at FDR <25% and nominal P<5% in PCS1 subtypes, including epithelial to mesenchymal transition, myogenesis, inflammatory response and so on. PCS2 and PCS3 both enriched in HALLMARK_MYC_TARGETS_V2 (a subgroup of genes regulated by MYC).
Discussion
In order to explore the prostate adenocarcinoma, we first studied RNA-Seq data of prostate tumor samples from TCGA network (n=497). The most robust and optimal number of classification was three identified by an unsupervised consensus-based clustering algorithm. We built up the PCS classifier (183 genes) based on the expression of encoded genes from RNA-Seq level, rather than those specialized genes filtered or corrected with clinical data. Those samples with same gene-expression pattern would be more likely to be identified and classified into the same category. Then the classifier was validated in three independent datasets, which were completely different in data platform, even in data type (two were mRNA microarray data, one was extron microarray data). From the results, we can see that the variation of genes would indicate diverse clinical outcome.
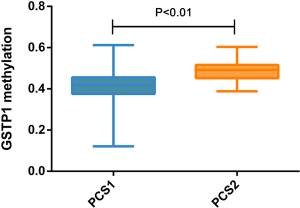
According to previous studies, tumorigenesis is closely related to internal or genetic disorders. These disorders sometimes can be observed by different gene expression, such as tumor suppressor gene mutations with reduced expression and oncogene activation with increased expression. Tumors need a continuous process. The patterns of disorder vary among different tumors. In other words, there is only one way to be normal, but many ways to be abnormal. These compose the basis of tumor subgrouping and precision medicine (34). Our three distinct prostate adenocarcinoma subtypes could be identified each associating with unique molecular features. For example, PCS1 was featured with highly expressed GSTP1 (P-class glutathione S-transferase gene 1) (P=9.62×10−68), which has been recognized as a tumor suppressor gene. Furthermore, there is a low hypermethylation of GSTP1 promoter in these PCS1 samples (P<0.01, Figure S1). Hypermethylation of the GSTP1 promoter is possibly the most common genetic event in prostate cancer and appears to be an early event in tumorigenesis (35). Usually, a low hypermethylation also means a high gene expression. Our observation corresponds to the fact that PCS1 patients have a relatively better prognosis. All the classifier genes (n=183) were functionally annotated with GO and KEGG databases. The most possible items were cell communication, development process and focal adhesion, most of which were closely related to tumor biology behavior.
We performed statistical test with matched TCGA clinical data among different groups, and found some interesting phenomena surprisingly. There were significant differences among these five Gleason grade groups (P<0.001). The same tendency was observed on pathology T stage and N stage, though we could not rule out the confounding effect here. We didn’t find any significant difference in recurrent time among three subtypes. Just as what the prostate adenocarcinoma global analysis said (12), the follow-up time in the TCGA project is insufficient in terms of those slow progression tumors such as prostate cancer. Besides, the follow-up rate of prostate adenocarcinoma dataset (less than 66%) and median follow-up time (15.1 months) might be additional reasons that alter the accuracy and credibility of the survival analysis.
For further validation, the same workflow was applied to another three GEO series. It showed significant differences among clusters in Gleason scores (P<0.05). In addition, Gleason grade group 2 (7=3+4) and Gleason grade group 3 (7=4+3) had obvious differences in GSE70769. Previous studies indicated the different prognosis between these two groups (36,37). Therefore, the new WHO grading system regarded them as 2 groups. Our result could show this difference, which further illustrated the subtle relationship between tumor subtypes and prognosis.
GSE70768 and GSE70769, which include more complete follow-up, have follow-up time of 27.5 and 81.1 months respectively. For GSE70769, although the difference of recurrences between PCS1 and PCS2 was near a critical position of statistical (P=0.059), PCS2 still has a tendency of bad prognosis. GSE70768 consisted of 125 samples, and the recurrence times between PCS1 and PCS2 was significantly different (P=0.005). We all know that the Gleason score is based on microscopic appearance. The morphology change may be based on the inherent genetic change of tumor cells. Is the tumor subtypes we obtain through PCS classifier the basis of architectural patterns? The positive results above, must mean something, which we should run after for a long time.
There are several prostate cancer classifications over the past years (26,38,39). Tomlins described 4 subtypes based on microarray gene expression patterns that are related to several genomic aberrations; You and colleges used pathway activation signatures of known relevance to prostate cancer to developed a novel classification system. Different from above, our PCS classifier mainly built on the mining of the whole expression profile by unsupervised clustering. Whether our model corresponds to the former reported classification, especially the basal and luminal subtypes? We analyzed the mean expression of basal and luminal markers among PCS subtypes. The results provide evidences that PCS1 tumors correspond to basal subtypes, PCS2 and PCS3 reflect luminal subtypes (Figure 5). PCS1 (basal like) subtypes have a better prognosis, the same as what You et al. reported. Aberrant CpG methylation hypermethylation of GSTP1 promoter mentioned above may participated in these biological processes. GSEA may give us some biological insights about 3 PCS subtypes. As a result, epithelial-to-mesenchymal transition gene sets are upregulated in PCS1 and significant at FDR <25% (Figure S2). Epithelial to mesenchymal transition, has been recognized as a feature of aggressive tumors, and plays a crucial importance in cancer invasion and metastasis (40,41). PCS2 and PCS3 both enriched in a gene sets regulated by MYC (Figures S3,S4). The MYC oncogene encodes a nuclear protein that is involved in the control of normal cell growth, differentiation, and apoptosis. Overexpression correlate with advancing stage and grade of prostate cancer (42-44).
There were still some limitations in our study. Only RNA-Seq data were concerned in subgrouping the tumors. In addition, we have some doubts about the meaning of PCS3, for the limited number especially in the validation datasets. Genetically, it was nearer to PCS2 (Figure 2B), though it had ambiguous outcome. Those above may implied the complexity of genes.
Conclusions
In conclusion, we explored the TCGA prostate adenocarcinoma database by machine learning. We built up the PCS classifier (183 genes) based on gene expression profile, and got three prostate adenocarcinoma subtypes. There were distinct differences among subgroups in Gleason grade group, pathological T, lymph node invasive and so on. These results were validated in other GEO datasets, and prognostic information also had significance in survival analysis, which might be helpful to further study and clinical use.
Full table

Full table

Full table
Full table
Acknowledgments
We would like to extend our gratitude to all staff working in TCGA network and the donors of tissues. We appreciate Desousa’s methods, which provided us with so much guidance.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.05.15). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This article does not contain any studies with human participants or animals performed by any of the authors. TCGA is a deidentified database. The access to the TCGA database is approved by the National Cancer Institute.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ketchandji M, Kuo YF, Shahinian VB, et al. Cause of death in older men after the diagnosis of prostate cancer. J Am Geriatr Soc 2009;57:24-30. [PubMed]
- Garnick MB. Prostate cancer: screening, diagnosis, and management. Ann Intern Med 1993;118:804-18. [Crossref] [PubMed]
- Walsh PC. Cancer surveillance series: interpreting trends in prostate cancer--part I: evidence of the effects of screening in recent prostate cancer incidence, mortality, and survival rates. J Urol 2000;163:364-5. [PubMed]
- Epstein JI, Egevad L, Amin MB, et al. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am J Surg Pathol 2016;40:244-52. [PubMed]
- Humphrey PA, Moch H, Cubilla AL, et al. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part B: Prostate and Bladder Tumours. Eur Urol 2016;70:106-19. [Crossref] [PubMed]
- van den Bergh RC, van der Kwast TH, de Jong J, et al. Validation of the novel International Society of Urological Pathology 2014 five-tier Gleason grade grouping: biochemical recurrence rates for 3+5 disease may be overestimated. BJU Int 2016;118:502-5. [Crossref] [PubMed]
- Qian J, Hirasawa K, Bostwick DG, et al. Loss of p53 and c-myc Overrepresentation in Stage T2-3N1-3M0 Prostate Cancer are Potential Markers for Cancer Progression. Mod Pathol 2002;15:35-44. [Crossref] [PubMed]
- Catz SD, Johnson JL. BCL-2 in prostate cancer: a minireview. Apoptosis 2003;8:29-37. [Crossref] [PubMed]
- Cancer Genome Atlas Research N. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113-20. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061-8. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609-15. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015;163:1011-25. [Crossref] [PubMed]
- De Sousa E, Melo F, Wang X, Jansen M, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med 2013;19:614-8. [Crossref] [PubMed]
- Wong KC, Li Y, Peng C, et al. Computational learning on specificity-determining residue-nucleotide interactions. Nucleic Acids Res 2015;43:10180-9. [PubMed]
- Erho N, Crisan A, Vergara IA, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. Plos One 2013;8:e66855 [Crossref] [PubMed]
- Ross-Adams H, Lamb AD, Dunning MJ, et al. Integration of copy number and transcriptomics provides risk stratification in prostate cancer: A discovery and validation cohort study. Ebiomedicine 2015;2:1133-44. [Crossref] [PubMed]
- Zhu Y, Qiu P, Ji Y. TCGA-Assembler: open-source software for retrieving and processing TCGA data. Nat Methods 2014;11:599-600. [Crossref] [PubMed]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572-3. [Crossref] [PubMed]
- Monti S, Tamayo P, Mesirov J, et al. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Mach Learn 2003;52:91-118. [Crossref]
- Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. J R Statist Soci 2001;63:411-23. [Crossref]
- Kaufman L, Rousseeuw PJ. Finding groups in data. an introduction to cluster analysis. Biometrics 1990;
- Rousseeuw PJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 1987;20:53-65. [Crossref]
- Schwender H, Ickstadt K. Empirical Bayes analysis of single nucleotide polymorphisms. BMC Bioinformatics 2008;9:144. [Crossref] [PubMed]
- Sing T, Sander O, Beerenwinkel N, et al. ROCR: visualizing classifier performance in R. Bioinformatics 2005;21:3940-1. [Crossref] [PubMed]
- Tibshirani R, Hastie T, Narasimhan B, et al. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A 2002;99:6567-72. [Crossref] [PubMed]
- You S, Knudsen BS, Erho N, et al. Integrated Classification of Prostate Cancer Reveals a Novel Luminal Subtype with Poor Outcome. Cancer Res 2016;76:4948-58. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Lee WH, Isaacs WB, Bova GS, et al. CG island methylation changes near the GSTP1 gene in prostatic carcinoma cells detected using the polymerase chain reaction: a new prostate cancer biomarker. Cancer Epidemiol Biomarkers Prev 1997;6:443-50. [PubMed]
- Brooks JD, Weinstein M, Lin X, et al. CG island methylation changes near the GSTP1 gene in prostatic intraepithelial neoplasia. Cancer Epidemiol Biomarkers Prev 1998;7:531-6. [PubMed]
- Palmer DH, Hussain SA, Ganesan R, et al. CD40 expression in prostate cancer: a potential diagnostic and therapeutic molecule. Oncol Rep 2004;12:679-82. [PubMed]
- McGrath MJ, Binge LC, Sriratana A, et al. Regulation of the transcriptional coactivator FHL2 licenses activation of the androgen receptor in castrate-resistant prostate cancer. Cancer Res 2013;73:5066-79. [Crossref] [PubMed]
- Sørensen KD, Abildgaard MO, Haldrup C, et al. Prognostic significance of aberrantly silenced ANPEP expression in prostate cancer. Br J Cancer 2013;108:420-8. [Crossref] [PubMed]
- Sun GG, Lu YF, Zhang J, et al. Filamin A regulates MMP-9 expression and suppresses prostate cancer cell migration and invasion. Tumour Biol 2014;35:3819-26. [Crossref] [PubMed]
- Fisher KW, Montironi R, Lopez Beltran A, et al. Molecular foundations for personalized therapy in prostate cancer. Curr Drug Targets 2015;16:103-14. [Crossref] [PubMed]
- Lee WH, Morton RA, Epstein JI, et al. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci U S A 1994;91:11733-7. [Crossref] [PubMed]
- Jo JK, Hong SK, Byun SS, et al. Comparison of clinical outcomes between upgraded pathologic Gleason score 3 + 4 and non-upgraded 3 + 4 prostate cancer among patients who are candidates for active surveillance. World J Urol 2015;33:1729-34. [Crossref] [PubMed]
- Koontz BF, Tsivian M, Mouraviev V, et al. Impact of primary Gleason grade on risk stratification for Gleason score 7 prostate cancers. Int J Radiat Oncol Biol Phys 2012;82:200-3. [Crossref] [PubMed]
- Smith BA, Sokolov A, Uzunangelov V, et al. A basal stem cell signature identifies aggressive prostate cancer phenotypes. Proc Natl Acad Sci U S A 2015;112:E6544-52. [Crossref] [PubMed]
- Tomlins SA, Alshalalfa M, Davicioni E, et al. Characterization of 1577 primary prostate cancers reveals novel biological and clinicopathologic insights into molecular subtypes. Eur Urol 2015;68:555-67. [Crossref] [PubMed]
- Thiery JP, Acloque H, Huang RYJ, et al. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139:871-90. [Crossref] [PubMed]
- Gravdal K, Halvorsen OJ, Haukaas SA, et al. A switch from E-cadherin to N-cadherin expression indicates epithelial to mesenchymal transition and is of strong and independent importance for the progress of prostate cancer. Clin Cancer Res 2007;13:7003-11. [Crossref] [PubMed]
- Buttyan R, Sawczuk IS, Benson MC, et al. Enhanced expression of the c-myc protooncogene in high-grade human prostate cancers. Prostate 1987;11:327-37. [Crossref] [PubMed]
- Jenkins RB, Qian J, Lieber MM, et al. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res 1997;57:524-31. [PubMed]
- Qian J, Hirasawa K, Bostwick DG, et al. Loss of p53 and c-myc overrepresentation in stage T(2-3)N(1-3)M(0) prostate cancer are potential markers for cancer progression. Mod Pathol 2002;15:35-44. [Crossref] [PubMed]