
Genetically engineered mouse models for studying radiation biology
Introduction
Radiation therapy is one of the three major modalities for treating cancer, along with surgery and chemotherapy. Nearly half of all cancer patients will receive radiation therapy in either a curative or palliative setting (1). Clinically, radiation oncologists and physicists have harnessed milestones in physics and engineering to improve outcomes for patients receiving radiation. Such milestones include the discovery of X-rays, protons, and nuclear magnetic resonance (NMR). These discoveries were often translated to various technologies to improve patient care, ranging from the development of linear accelerators in the 1950s, to computed tomography (CT) and magnetic resonance imaging (MRI) in the 1970s, to the application of advanced radiation field-shaping collimators and imaging for 3D treatment planning in the 1980–1990s (2). Similar scientific milestones have been reached in radiobiology, and translating these advances to improve clinical outcomes by harnessing biological mechanisms remains a critical area of current research. GEMMs can serve as powerful preclinical models to accelerate this translation.
Suitable systems for modeling cancer in vivo should strive to satisfy several key features, particularly for radiobiology research. First, the tumor model should mimic human cancers on a histological, anatomical, and genetic level. Tumor models should be initiated by the same driver mutations observed in human tumors of the same type being studied. Further, the animal tumors should resemble human tumors anatomically and histologically with classic histopathological features used for diagnosis. Second, the tumor microenvironment should be preserved as best as possible. The importance of the microenvironment has been demonstrated most strikingly in studies showing that tumor vasculature and hypoxia in transplanted models differ from spontaneous models (3-5). This consideration is especially relevant for studying how the response to ionizing radiation is impacted by the tumor microenvironment and immune system (6,7). Third, ideally tumor initiation should be controlled spatially, temporally, and genetically. Precise control over tumor initiation is important for experimental reproducibility and feasibility. Most importantly, because cancer is a genetic disease, models should enable facile gene manipulation for systematic dissection of normal and tumor tissue response to radiation. Xenograft models are relatively simple to implement in a research pipeline and remain a workhorse model in research and industry (8). However, xenografts only satisfy some of the ideal features of a model system to study tumor biology, and it is therefore not surprising that xenografts may lack predictive value for clinical outcomes (5,8,9). In contrast, GEMMs provide many if not all of these features. Thus, GEMMs enable creative, rigorous, and reproducible experimental designs for studying radiobiology.
Current modeling techniques allow scientists to manipulate genes in several ways: activation, knockdown, knockout, tissue-specific expression, inducible expression, and sequential expression. Here, we introduce these GEMM technologies through the lens of radiobiology in three contexts: (I) studying normal tissue radiation injury; (II) dissecting tumor response to radiation therapy; (III) developing next-generation mouse models.
In vivo short hairpin RNA (shRNA) to investigate normal tissue radiation injury and carcinogenesis
As the number of people diagnosed with cancer each year grows, so does the number of patients living with the long-term complications of radiation therapy. Approximately half of all patients diagnosed with cancer will receive radiation as a part of their therapy regimen (10,11). Regardless of whether exposure to ionizing radiation is the result of medical therapy or accidental exposure, normal healthy tissues are inadvertently exposed to radiation, which may result in a variety of toxicities and even secondary malignancies. Despite the rapid advances in radiation oncology that reduce exposure of surrounding healthy tissues by enabling radiation to be delivered precisely to tumor sites, collateral exposure of normal tissue is unavoidable. As the number of radiation survivors increases, it is essential to understand the mechanisms regulating normal tissue radiation injury and radiation carcinogenesis.
In some radiosensitive tissues, such as the hematopoietic system, the tumor suppressor p53 promotes the apoptotic cell death and acute tissue toxicity associated with radiotherapy (12-14). Therefore, blocking p53-mediated cell death during radiotherapy may represent an effective treatment strategy for cancer patients by minimizing the injury to normal tissue without obstructing the response of p53 mutant tumors (14-16). In order to model the temporary loss of p53 that would occur with a p53 inhibitor, we utilized the tetracycline-responsive element (TRE) to drive the in vivo expression of a shRNA against p53 (17,18). The reverse tetracycline-controlled transactivator (rtTA) was expressed ubiquitously and induced expression of the p53 shRNA only in the presence of doxycycline (Figure 1). These compound transgenic mice enabled us to decrease p53 mRNA expression in an inducible and reversible manner in p53 wild type mice. Temporary induction of the p53 shRNA during total body irradiation improved the survival of mice from the hematopoietic acute radiation syndrome by protecting hematopoietic stem and progenitor cells (HSPC) from radiation-induced cell death. These results suggest that a p53 inhibitor given concurrently with radiotherapy may ameliorate radiation-induced injury in the hematopoietic system.
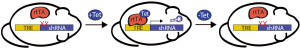
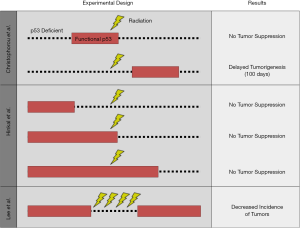
Although temporary knockdown of p53 with this tetracycline-regulated mouse model protected normal tissue from radiation, mice with germline p53 deletion develop radiation-induced cancer (19,20). Therefore, one potential risk associated with p53 inhibition during radiotherapy is that the incidence of secondary tumors may be exacerbated. Other researchers evaluated the tumor suppressor function of p53 during irradiation, but these studies made use of mice that were functionally p53 null or had permanent deletion of p53 (Figure 2) (21,22). One key advantage of the tetracycline-controlled p53 shRNA mouse model is that p53 expression is intact in the absence of doxycycline, mimicking the wild type p53 status of most cancer patients. This model was therefore well-equipped to address the question: does temporary loss of p53 during radiation increase the incidence of radiation-induced lymphomas? Surprisingly, we found that temporarily blocking p53 during fractionated radiation exposure enhanced the overall survival of mice by preventing lymphomagenesis. Knockdown of p53 during radiation also promoted the regeneration of the hematopoietic system by enhancing the fitness of HSPCs within the bone marrow. Furthermore, transplantation of bone marrow cells from mice with temporary loss of p53 during radiation into irradiated recipients was sufficient to suppress lymphomagenesis. Together, these data revealed a new paradigm for the role of p53 during radiation: within the bone marrow, p53 promotes radiation-induced lymphomagenesis in a non-cell-autonomous manner.
Cre-loxP technology to study normal tissue radiation injury
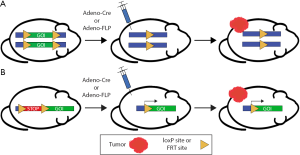
The risk of late effects from radiation therapy frequently limits the maximum radiation dose that can be safely delivered to a malignancy. Therefore, studies to understand the mechanisms of radiation-induced normal tissue injury may be translated to increase the radiation dose to a tumor while preventing normal tissue injury. Cre-loxP technology is well-suited to dissect mechanisms of radiation-induced normal tissue injury. The bacteriophage P1-derived Cre-loxP system can be used to carry out site-specific deletions, insertions, and translocations within genomic DNA (Figure 3). The enzyme Cre recombinase recombines a pair of short palindromic sequences called loxP sites, which are inserted into genomic DNA to flank a target (‘floxed’) locus. Activity of the Cre recombinase can be controlled by various mechanisms, including ubiquitous expression, expression under a cell type-specific promoter, use of a ligand-inducible system (such as tamoxifen), or viral packaging for targeted injection and delivery. We used Cre-loxP technology to delete p53 specifically in endothelial cells using the Tie2-Cre and VE-Cadherin-Cre mice to demonstrate the role of endothelial cell-specific p53 in preventing radiation-induced myocardial injury in mice (23). Radiation-related heart disease is an important cause of treatment-associated mortality for patients receiving thoracic radiotherapy (24), and the findings from this preclinical work suggest that combining radiation therapy with inhibitors of p53 (25) may increase the risk for cardiac injury if the heart is in the radiation field.
However, the role of Cre-loxP technology goes beyond modeling and understanding the mechanisms of radiation-related normal tissue injury. These GEMMs can also be used to study biomarkers of normal tissue injury as well as various imaging modalities that can potentially be translated for early clinical intervention. For example, the Tie2-Cre; p53fl/fl mouse model has several characteristics that make it useful to study radiation-induced heart disease: genetic manipulation leads to decreased latency of disease compared to irradiation of wild type mice, and the severity of the cardiomyopathy is such that mortality reaches 100%, as opposed to 50% in genetically intact mice (26). Furthermore, the dose of radiation delivered in a single fraction to the heart to elicit such a profound phenotype is only 60% of that delivered to wild type mice and rats (12 vs. 20 Gy) (27). This Tie2-Cre; p53fl/fl model utilizes Cre-loxP technology to exploit the biology of the DNA damage response to generate an efficient model of radiation-induced normal tissue injury with high penetrance, accelerated onset, and a severe phenotype. This GEMM has also been adopted for the study of dual energy-microCT, 4D-microCT, and microSPECT imaging to assess changes in vascular permeability and cardiac function (28), which are surrogates for myocardial injury. These preclinical imaging approaches developed in GEMMs have the potential to be translated as imaging biomarkers to identify patients treated with radiation who have developed cardiac injury that may progress to radiation-related heart disease.
In addition to the development of preclinical imaging modalities using GEMMs, Cre-mediated recombination of fluorescent reporters can also be used for additional resolution of cellular proliferation and lineage tracing. Cre-mediated recombination of the confetti allele can be used to randomly tag airway progenitor cells with one of four fluorescent lineage reporters. Using this technique to perform in vivo lineage tracing, Farin et al. revealed the stark difference between the effects of low- and high-linear energy transfer (LET) radiation in vivo and in vitro. While radiation induces a decrease in colony forming ability in vitro, a significant increase in clonal expansion of progenitor cells is observed in vivo following exposure to either low- or high-LET radiation (29). These findings suggest that a reduction in proliferation-competent progenitor cells after radiation exposure actually results in clonal expansion of the remaining progenitor cells in vivo. This example demonstrates the application of Cre-loxP technology for lineage tracing of specific progenitor cells for in vivo clonogenic assays, which exhibit dramatically different results compared to those performed using traditional in vitro cell culture methods.
Concern over the potential exposure of civilians to high-dose radiation has led to increased interest in understanding mechanisms underlying radiation injury. Specifically, no medical countermeasures are approved to prevent or treat the radiation-induced gastrointestinal (GI) syndrome, and the cellular targets of radiation-induced toxicity in the GI tract and molecular mechanisms underlying cell death in the GI syndrome remain active areas of investigation. Using Cre-loxP technology to delete p53 and essential mediators of the intrinsic apoptotic cascade specifically in GI epithelial cells, we found that p53 controlled the radiation-induced GI syndrome in mice through a mechanism independent of apoptosis (30). These experiments utilize genetic manipulation within a particular cellular compartment to elucidate a specific mechanism for acute radiation syndrome. Studies such as these have significant implications for efforts to develop medical countermeasures against radiation due to the cellular and molecular resolution they provide.
Cre-loxP and FLP-FRT technology to dissect mechanisms of tumor response to radiation
Cre recombinase has been widely utilized for the simultaneous manipulation of multiple genes to study tumorigenesis and tumor progression in the mouse model system, leading to the development of GEMMs of a wide variety of primary cancers. As described above, the activity of the site-specific recombinase Cre, as well as an analagous recombinase derived from Saccharomyces cerevisiae called flippase (FLP), can be controlled by various mechanisms, allowing ubiquitous, cell type-specific, or inducible activity (Figure 3). Through these various mechanisms, one can spatially and temporally restrict genetic recombination by Cre and FLP at loxP and FLP recombinase target (FRT) sites (respectively) in order to investigate mechanisms of tumor formation, progression, and response to radiation therapy.
These tools have proven to be particularly well-suited for the study of radiation biology. For example, use of Cre-loxP technology to generate GEMMs of primary non-small cell lung cancer (NSCLC) with various initiating mutations has revealed that genetic factors within histologically indistinguishable tumor parenchymal cells influence in vivo tumor growth delay after radiation (31), which has important clinical implications for patients receiving radiation therapy. Furthermore, the use of micro-CT has been adopted in order to better visualize and monitor the response of primary murine lung tumors to radiation, demonstrating the ability of advanced imaging technology to impact the field of radiation biology (32). In addition, we have employed Cre-loxP technology to investigate the role of hypoxia-inducible factor 1-alpha (HIF-1α) and hypoxia in mediating tumor response to radiation therapy (33).
The efficiency and relative simplicity of Cre-loxP technology have led to its widespread use to study cancer and radiation biology. Because most primary GEMMs of cancer utilize Cre-loxP technology to initiate carcinogenesis, the utility of Cre recombinase in subsequent modification of either tumor or stromal cells is limited (Figure 4A). In contrast to the growing number of floxed alleles and tissue-specific Cre drivers, the FLP-FRT system has been utilized less frequently than the Cre-loxP system to modify genes in somatic tissues in mice. The limitation of these single recombinase models is that they are often used to generate the driver mutations for tumorigenesis, but they are limited in the ability to ask more complex questions regarding the mechanisms of radiation response and radiation resistance. For example, the response of tumors to radiation therapy is mediated by a complex interaction between immune cells, stroma, and tumor cells. GEMMs present a unique opportunity to dissect the role of specific cellular compartments in tumor response to radiation therapy. However, to harness the full potential of site-specific recombinase technology to dissect mechanisms of radiation response, more than one recombinase is needed to not only initiate tumorigenesis but also to manipulate other cellular compartments to examine their role in the radiation response. Therefore, utilization of additional FRT-flanked alleles (‘FRTed’) alleles in concert with floxed alleles enables dual recombinase technology to direct distinct gene mutations to different cell types when Cre and FLP recombinases are employed simultaneously (Figure 4B).
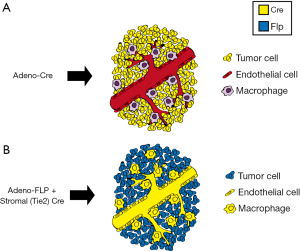
We have utilized this dual recombinase (Cre + FLP) technology to address a critical question in radiation biology: defining the cellular target of radiation therapy in tumor eradication (7). To investigate whether stromal cells, such as endothelial cells, are critical targets of radiation therapy, we applied dual recombinase technology. We generated multiple murine tumors which were genetically identical except for the deletion of the DNA damage response gene ataxia telangiectasia mutated (Atm). By deleting Atm in either tumor cells or endothelial cells, we were able to radiosensitize distinct cellular populations to better understand their roles in tumor response and eradication following radiation therapy. In these experiments, an adenovirus expressing FLP recombinase (Adeno-FLP) was used to initiate sarcoma development in the lower extremity. Adeno-FLP was injected intramuscularly to delete both copies of a conditional FRTed allele of p53 (p53FRT/FRT) (Figure 3A) (34). In addition, FLP excises a FRTed STOP cassette (FSF) upstream of oncogenic Kras to initiate transcription of KrasG12D (Figure 3B). In order to manipulate the radiosensitivity of endothelial cells, an endothelial-specific Cre driver (VE-Cadherin) recombines floxed alleles specifically in endothelial cells. In this case, Atm was selectively deleted in endothelial cells to enhance their radiosensitivity. We used this dual recombinase technology in KrasFSF-G12D; p53FRT/FRT; VE-Cadherin-Cre; Atmfl/fl mice (Table 1) in order to demonstrate that selectively sensitizing endothelial cells to mitotic cell death by deleting Atm (35) prolongs sarcoma growth delay after stereotactic body radiation therapy (SBRT) (36), but this enhanced radiosensitivity does not translate into improved local control (7). However, radiosensitization of tumor cells by deleting Atm increases local control of primary sarcomas after radiation therapy, demonstrating that tumor cells but not endothelial cells are critical targets of curative radiation therapy (7). These results have important clinical implications. They suggest that the increased local control that is observed with some tumors following SBRT may not be due to increased endothelial cell death, but is instead a consequence of delivering larger (10 to 20 Gy per fraction), biologically effective doses that kill more tumor cells compared to standard (1.8 to 2 Gy per fraction) radiation therapy (7).

Full table
By coupling the Cre-loxP and FLP-FRT systems, dual recombinase technology has made it possible to genetically manipulate both tumor cells and stromal cells (Figure 4) to address the role of various stromal cell populations in mediating response to radiotherapy. This capability gains increasing importance in the field of radiation biology because of the potential role of manipulating the immune system during radiotherapy to not only optimize local control, but also to potentially cause abscopal responses in distant sites of metastatic disease.
RCAS-TVA system to model primary cancers
Although genetic engineering in mice through the use of Cre-loxP technology has facilitated numerous advances in the field of radiation biology, these studies are often expensive and time-consuming. In vivo retroviral gene transfer represents a practical alternative for lineage tracing and cell fate mapping, generating mouse models of cancer, and evaluating the function of genes that regulate radiation response. The stable delivery of genes to somatic cells has been optimized using the RCAS (replication-competent avian sarcoma-leukosis virus long-terminal repeat with splice acceptor) viral vector, which was derived from the Rous sarcoma virus-A. In order to generate the RCAS vector, the oncogenic v-src was replaced with a multiple cloning site to accommodate genes of interest (37). The RCAS virus maintains expression of the subgroup A envelope glycoprotein and can therefore infect only cells with the cognate tumor virus A (TVA) receptor. Although the TVA receptor is naturally expressed only in avian cells, a host of genetically engineered mice expressing TVA under the control of a variety of tissue-specific promoters has been generated (38). Either virus-producing avian cells or the virus itself can be injected into transgenic mice that harbor the TVA receptor in order to manipulate gene expression in a spatiotemporal manner (Figure 5). The RCAS-TVA system can also be utilized in combination with Cre-loxP technology or with the tetracycline-controlled transcriptional regulation system to provide a more intricate level of control over gene transfer.
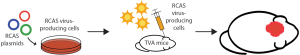
The RCAS-TVA system has been widely utilized to generate mouse models of cancer due to the ease through which mutant genes can be expressed and oncogenes can be stably overexpressed in a tissue-specific manner. Furthermore, mammalian cells that have been infected by an RCAS virus remain susceptible to reinfection, enabling the simultaneous or sequential manipulation of several genes. Therefore, the RCAS-TVA system has made it possible to model the complex mutational landscape and the clonal evolution of primary tumors.
The first models of cancer using the TVA receptor-mediated delivery of RCAS were developed by targeting glial cells within the mouse brain (39-41). These original models of gliomagenesis have been adapted over the last two decades to reflect the newly discovered landscape of genetic alterations that occur in human brain tumors (42). For example, a recent study utilized an RCAS construct carrying platelet derived growth factor-B (PDGFB) to drive the initiation of brainstem gliomas in mice expressing TVA under the control of the glial precursor cell-specific Nestin promoter (43). To more effectively model the alterations commonly observed in cell cycle regulatory genes in patients with brainstem gliomas, this Nestin-TVA mouse was crossed to an Ink4a/ARF null strain. The resulting GEMM was then utilized in a preclinical trial to evaluate the efficacy of inhibiting the cyclin/cyclin-dependent kinase/retinoblastoma signaling pathway in combination with radiotherapy (43). Treatment with a cyclin-dependent kinase 4/6 inhibitor following whole brain irradiation induced cell cycle arrest and significantly prolonged the survival of mice with Ink4a/ARF deficient brainstem gliomas. The effectiveness of the CDK4/6 inhibitor in this preclinical study also led to the initiation of a clinical trial for pediatric patients with brainstem gliomas (NCT02255461). The RCAS-TVA system for mouse modeling has enabled many other preclinical trials to test drugs that improve the effectiveness of radiation therapy. Gemcitabine, a drug that inhibits the synthesis of nucleic acids, significantly extends survival in an Ink4a/ARF and PTEN-deficient mouse model of glioblastoma when administered concurrently with radiation (44). In this same model, concomitant treatment with a poly (ADP-ribose) polymerase (PARP) inhibitor, temozolomide, and radiotherapy led to an improved tumor growth delay and overall survival when compared with temozolomide and radiation alone, the current standard-of-care (45).
In addition to facilitating preclinical trials, the RCAS-TVA system has been elegantly used to begin dissecting the mechanisms of radioresistance in gliomas. In primary models of medulloblastoma and glioblastoma, researchers identified the nucleus dense, bulk tumor region as preferentially undergoing apoptosis in response to radiotherapy, while cells in the perivascular regions appeared more radioresistant (46,47). Brain tumor stem cells and supporting stromal cell populations have been shown to reside in the perivascular niche, highlighting the need for autochthonous tumor models when evaluating the radiation response of tumors. Hambardzumyan et al. went on to show that perivascular cells staining positively for various stem cell markers underwent p53-mediated cell cycle arrest following radiation. This cancer stem cell population ultimately survived the radiotherapy and re-entered the cell cycle 72 hours post radiation. Additional mechanistic insight into the radioresistant nature of gliomas was elucidated by studying the defects in the DNA damage response pathway commonly found in human tumors. The genetic loss of Chk2 in a mouse model of glioblastoma, for example, protected tumor cells from radiation-induced apoptosis and eradicated the survival benefit following radiotherapy (48). Additional studies are required to determine whether the role of Chk2 in the radioresistance of glioblastomas can be generalized to other components of the DNA damage response pathway.
Next-generation mouse modeling of cancer with CRISPR/Cas9 technology
The research described so far demonstrates the broad utility of GEMMs in radiobiology. Despite the immense potential for GEMMs in radiation research, the time and financial costs for model generation and execution of adequately powered experiments can be prohibitive. The conventional approach for mouse model generation relies on inserting targeted donor constructs via homologous recombination (HR) in embryonic stem (ES) cells, targeted ES cell selection and injection into blastocysts, implantation, and months of breeding (49). This entire process of generating germline chimeras can take several months before mice with the desired genotype are in hand. If more than one gene modification is desired, then the time frame increases by several more months of extensive breeding.
Recently, both the time and financial costs associated with generating mouse models have drastically decreased, especially for mouse models of cancer. The discovery and engineering of a bacterial adaptive immune system that utilizes clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) have made this possible (50-52). CRISPR/Cas is an RNA-guided endonuclease system that can be distilled to three basic components as used for genome editing in mammalian cells: (I) a Cas protein; (II) a single guide RNA (sgRNA); and (III) a protospacer adjacent motif (PAM) (Figure 6). The Cas proteins, of which Cas9 is the most widely used, serve as molecular scissors that make double strand breaks (DSBs) in DNA. Cas9 is targeted to DNA by a chimeric sgRNA through sequence complementarity between the sgRNA and the genomic DNA. The endonuclease activity of Cas9 is dependent on the presence of a PAM sequence adjacent to the 3' end of the sgRNA target sequence (53).
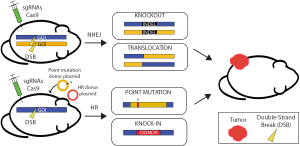
CRISPR/Cas technology can help bypass the two primary bottlenecks during generation of cancer GEMMs: (I) targeted insertion in ES cells by HR and (II) breeding of progeny. The fundamental basis for all gene editing technology is the repair of DSBs in DNA. Prior to CRISPR/Cas, several methods were used for generating targeted DSBs including site-specific endonucleases, zinc-finger nucleases (ZFNs), and transcription activator-like effector nucleases (TALENs) (54,55). These nucleases represented an evolution in improving DNA targeting specificity and flexibility. However, prior to CRISPR/Cas, intensive protein engineering was required to utilize ZFN and TALEN technology because cleavage specificity was protein-dependent. As such, ZFNs and TALENs could achieve target specificity, but were not flexible because each new genomic target required engineering a new protein with DNA sequence specificity. In contrast, CRISPR/Cas is a programmable RNA-guided endonuclease, which allows for both specificity and flexibility of DNA targeting. Therefore, generating specific genome-wide DSBs for genome engineering became a relatively simple process of designing a new sgRNA with complementarity to the targeted DNA sequence in the genome. As importantly, Mali et al. discovered CRISPR/Cas9 had greater HR efficiency in mammalian cells compared to previous endonucleases when targeting the same locus (51). Thus, CRISPR/Cas can eliminate the HR bottleneck in GEMM generation. The second bottleneck occurs when chimeric mice must be crossed for several weeks to months to generate the desired genotype that may contain multiple alleles for tumor initiation. Because cancer most often arises stochastically in somatic tissues, if somatic gene edits could be generated in adult mice with CRISPR/Cas9 then this second bottleneck could be bypassed as well. In 2014, several labs demonstrated the successful generation of multiple non-germline GEMMs of cancer using CRISPR/Cas9 technology, thus completely bypassing the time-consuming process of mouse breeding.
The first groups to generate GEMMs using CRISPR/Cas9 successfully used in vivo somatic genome engineering to model several cancers in mice (56-59). These groups used CRISPR/Cas9 and mutagenic non-homologous end joining (NHEJ) repair to knockout tumor suppressor genes in the lung (56,59) and liver (57). Several groups even used CRISPR/Cas9 to generate the Eml4-Alk translocation to form lung tumors (58,60). Since these initial non-germline GEMMs, cancers of the breast (61), pancreas (62,63), brain (64), and leukemias (65) have also been modeled using CRISPR/Cas9. The generation of a Cas9 knock-in mouse has further streamlined non-germline GEMM development (59). These models resulted from the application of only the first widely used Cas protein, Cas9, to knock out tumor suppressor genes, or knock in oncogenes, and represented a leap forward in cancer mouse modeling. Using CRISPR/Cas9 in this way, the mouse genome can be modified in any way imaginable from simple point mutations to introducing genetic markers and chromosomal rearrangements (Figure 6).
CRISPR has been transformative for mouse modeling, but there are important limitations to consider. First, the targeting of Cas9 and other Cas proteins have unique PAM sequences that are required for targeting of the endonucleases. These sequence motifs are important for specificity, but may limit gene editing to certain areas of the genome. Fortunately, engineered variants of Cas9 and the discovery of novel Cas proteins with altered PAM specificities are minimizing the impact of this limitation (66). Second, off-target effects are important to consider when using CRISPR technology. Off-target effects resulting from small molecules have half-lives and the effect is temporary; in contrast, gene edits are permanent. Researchers have been extremely cognizant of this caveat to employing Cas9 technology since its inception (53,67-69). As such, several approaches to minimize the risk of these off-target effects have been developed. These approaches include the development of nickases that require two single-strand nicks to generate DSBs (70), high-fidelity Cas9 enzymes that have very few to no detectable off-target effects (71,72), and methods of detecting off-target mutations generated by Cas9Jeny (73-75).
Recently, the CRISPR/Cas9 system has evolved beyond simple genome editing into the realms of epigenome editing (76,77), base-pair editing (78), and genome regulation (79-81). Completely new classes of Cas proteins have also been discovered with altered PAM specificity that generate staggered DSBs (82), or Cas proteins that allow RNA-guided RNA targeting (83,84). These technologies expand the CRISPR toolbox for radiation biologists and enable truly next-generation mouse modeling and discoveries. The research performed to study acute and chronic radiation effects on normal tissue, radiation-induced carcinogenesis, and tumor response to radiation therapy using conventional GEMMs can potentially be accelerated at lower cost with CRISPR technology. For example, with CRISPR it is now feasible to study pathways activated by radiation such as the DNA damage response and apoptosis pathways in a functional way. Similar experiments using CRISPR have been performed to investigate metastasis and cooperative mutations required for tumorigenesis (65). CRISPR screens can also be performed to identify radioresistant and radiosensitizing genetic and epigenetic components, similar to screens performed with shRNA in the past (85-87).
Conclusions
Here we reviewed the applications of mouse models in preclinical radiobiology research. We introduced fundamental concepts of mouse modeling and summarized several technologies for mouse modeling including site-specific recombinases, shRNA, RCAS-TVA, and CRISPR. We demonstrated the utility of these technologies and mouse modeling in radiobiology research by highlighting several publications that used GEMMs in preclinical research on radiation-induced carcinogenesis, normal tissue injury, and the in vivo targets of radiation in tumors. Despite the important contributions GEMMs have made in radiation research, these and next-generation mouse models are poised to reveal additional insights in the future. Experiments performed in robust in vivo systems, such as GEMMs, will continue to lead to fundamental discoveries in radiation research. GEMMs of cancer are powerful tools for investigating the cell-autonomous and non-cell-autonomous mechanisms that contribute to normal and tumor tissue response to radiation. We anticipate that increased adoption of GEMMs in radiobiology research will not only lead to important new knowledge, but may also increase the predictive value of preclinical research to accelerate clinical translation and improve outcomes for patients treated with radiotherapy.
Acknowledgments
Funding: We acknowledge support from National Institutes of Health award R35 CA197616 to DG Kirsch, F30CA206424 to M Chen, and T32GM007171 to M Chen and AJ Wisdom.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Marco Durante, Giusi I. Forte, Giorgio Russo) for the series “Radiobiological models towards a personalized radiation oncology” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: DG Kirsch is a member of the scientific advisory board and owns stock in Lumicell Diagnostics, a company commercializing intraoperative imaging systems. DG Kirsch is a founder and owns stock in XRAD Therapeutics, which is developing radiosensitizers. DG Kirsch has research support from XRAD Therapeutics and Merck and has received research support in the past from GlaxoSmithKline and Janssen.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Moding EJ, Kastan MB, Kirsch DG. Strategies for optimizing the response of cancer and normal tissues to radiation. Nat Rev Drug Discov 2013;12:526-42. [Crossref] [PubMed]
- Bortfeld T, Jeraj R. The physical basis and future of radiation therapy. Br J Radiol 2011;84:485-98. [Crossref] [PubMed]
- Fenton BM, Lord EM, Paoni SF. Intravascular HbO2 saturations, perfusion and hypoxia in spontaneous and transplanted tumor models. Int J Cancer 2001;93:693-8. [Crossref] [PubMed]
- Field SB, Needham S, Burney IA, et al. Differences in vascular response between primary and transplanted tumours. Br J Cancer 1991;63:723-6. [Crossref] [PubMed]
- Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov 2006;5:741-54. [Crossref] [PubMed]
- Formenti SC, Demaria S. Systemic effects of local radiotherapy. Lancet Oncol 2009;10:718-26. [Crossref] [PubMed]
- Moding EJ, Castle KD, Perez BA, et al. Tumor cells, but not endothelial cells, mediate eradication of primary sarcomas by stereotactic body radiation therapy. Sci Transl Med 2015;7:278ra34 [Crossref] [PubMed]
- Singh M, Johnson L. Using genetically engineered mouse models of cancer to aid drug development: an industry perspective. Clin Cancer Res 2006;12:5312-28. [Crossref] [PubMed]
- Becher OJ, Holland EC. Genetically engineered models have advantages over xenografts for preclinical studies. Cancer Res 2006;66:3355-8, discussion 3358-9. [Crossref] [PubMed]
- Howlader N, Noone AM, Krapcho M, et al. SEER cancer statistics review, 1975-2012 [Internet]. Bethesda, MD: National Cancer Institute; 2013. [cited 2015 Jan 28].
- Ringborg U, Bergqvist D, Brorsson B, et al. The Swedish Council on Technology Assessment in Health Care (SBU) systematic overview of radiotherapy for cancer including a prospective survey of radiotherapy practice in Sweden 2001--summary and conclusions. Acta Oncol 2003;42:357-65. [Crossref] [PubMed]
- Pant V, Quintás-Cardama A, Lozano G. The p53 pathway in hematopoiesis: lessons from mouse models, implications for humans. Blood 2012;120:5118-27. [Crossref] [PubMed]
- Gudkov AV, Komarova EA. The role of p53 in determining sensitivity to radiotherapy. Nat Rev Cancer 2003;3:117-29. [Crossref] [PubMed]
- Gudkov AV, Komarova EA. Pathologies associated with the p53 response. Cold Spring Harb Perspect Biol 2010;2:a001180 [Crossref] [PubMed]
- Leonova KI, Shneyder J, Antoch MP, et al. A small molecule inhibitor of p53 stimulates amplification of hematopoietic stem cells but does not promote tumor development in mice. Cell Cycle 2010;9:1434-43. [Crossref] [PubMed]
- Su H, Ganapathy S, Li X, et al. p53-Based Strategy for Protection of Bone Marrow From Y-90 Ibritumomab Tiuxetan. Int J Radiat Oncol Biol Phys 2015;92:1116-22. [Crossref] [PubMed]
- Dickins RA, McJunkin K, Hernando E, et al. Tissue-specific and reversible RNA interference in transgenic mice. Nat Genet 2007;39:914-21. [Crossref] [PubMed]
- Lee CL, Castle KD, Moding EJ, et al. Acute DNA damage activates the tumour suppressor p53 to promote radiation-induced lymphoma. Nat Commun 2015;6:8477. [Crossref] [PubMed]
- Kemp CJ, Wheldon T, Balmain A. p53-deficient mice are extremely susceptible to radiation-induced tumorigenesis. Nat Genet 1994;8:66-9. [Crossref] [PubMed]
- Lee JM, Abrahamson JL, Kandel R, et al. Susceptibility to radiation-carcinogenesis and accumulation of chromosomal breakage in p53 deficient mice. Oncogene 1994;9:3731-6. [PubMed]
- Christophorou MA, Ringshausen I, Finch AJ, et al. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature 2006;443:214-7. [Crossref] [PubMed]
- Hinkal G, Parikh N, Donehower LA. Timed somatic deletion of p53 in mice reveals age-associated differences in tumor progression. PLoS One 2009;4:e6654 [Crossref] [PubMed]
- Lee C-L, Moding EJ, Cuneo KC, et al. p53 functions in endothelial cells to prevent radiation-induced myocardial injury in mice. Sci Signal 2012;5:ra52. [Crossref] [PubMed]
- Darby SC, Cutter DJ, Boerma M, et al. Radiation-related heart disease: current knowledge and future prospects. Int J Radiat Oncol Biol Phys. 2010;76:656-65. [Crossref] [PubMed]
- Gudkov AV, Komarova EA. Prospective therapeutic applications of p53 inhibitors. Biochem Biophys Res Commun 2005;331:726-36. [Crossref] [PubMed]
- Mezzaroma E, Di X, Graves P, et al. A mouse model of radiation-induced cardiomyopathy. Int J Cardiol 2012;156:231-3. [Crossref] [PubMed]
- Schultz-Hector S. Radiation-induced heart disease: review of experimental data on dose response and pathogenesis. Int J Radiat Biol 1992;61:149-60. [Crossref] [PubMed]
- Lee CL, Min H, Befera N, et al. Assessing cardiac injury in mice with dual energy-microCT, 4D-microCT, and microSPECT imaging after partial heart irradiation. Int J Radiat Oncol Biol Phys 2014;88:686-93. [Crossref] [PubMed]
- Farin AM, Manzo ND, Kirsch DG, et al. Low- and high-LET radiation drives clonal expansion of lung progenitor cells in vivo. Radiat Res 2015;183:124-32. [Crossref] [PubMed]
- Kirsch DG, Santiago PM, di Tomaso E, et al. p53 controls radiation-induced gastrointestinal syndrome in mice independent of apoptosis. Science 2010;327:593-6. [Crossref] [PubMed]
- Perez BA, Ghafoori AP, Lee CL, et al. Assessing the radiation response of lung cancer with different gene mutations using genetically engineered mice. Front Oncol 2013;3:72. [Crossref] [PubMed]
- Kirsch DG, Grimm J, Guimaraes AR, et al. Imaging primary lung cancers in mice to study radiation biology. Int J Radiat Oncol Biol Phys 2010;76:973-7. [Crossref] [PubMed]
- Zhang M, Qiu Q, Li Z, et al. HIF-1 Alpha Regulates the Response of Primary Sarcomas to Radiation Therapy through a Cell Autonomous Mechanism. Radiat Res 2015;183:594-609. [Crossref] [PubMed]
- Lee CL, Moding EJ, Huang X, et al. Generation of primary tumors with Flp recombinase in FRT-flanked p53 mice. Dis Model Mech 2012;5:397-402. [Crossref] [PubMed]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer 2003;3:155-68. [Crossref] [PubMed]
- Moding EJ, Lee CL, Castle KD, et al. ATM deletion with dual recombinase technology preferentially radiosensitizes tumor endothelium. J Clin Invest 2014;124:3325-38. [Crossref] [PubMed]
- Hughes SH, Greenhouse JJ, Petropoulos CJ, et al. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J Virol 1987;61:3004-12. [PubMed]
- von Werder A, Seidler B, Schmid RM, et al. Production of avian retroviruses and tissue-specific somatic retroviral gene transfer in vivo using the RCAS/TVA system. Nat Protoc 2012;7:1167-83. [Crossref] [PubMed]
- Holland EC, Varmus HE. Basic fibroblast growth factor induces cell migration and proliferation after glia-specific gene transfer in mice. Available online: http://www.pnas.org/content/95/3/1218.full.pdf
- Holland EC, Hively WP, DePinho RA, et al. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev 1998;12:3675-85. [Crossref] [PubMed]
- Holland EC, Celestino J, Dai C, et al. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet 2000;25:55-7. [Crossref] [PubMed]
- Wu G, Diaz AK, Paugh BS, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 2014;46:444-50. [Crossref] [PubMed]
- Barton KL, Misuraca K, Cordero F, et al. PD-0332991, a CDK4/6 inhibitor, significantly prolongs survival in a genetically engineered mouse model of brainstem glioma. PLoS One 2013;8:e77639 [Crossref] [PubMed]
- Galbán S, Lemasson B, Williams TM, et al. DW-MRI as a Biomarker to Compare Therapeutic Outcomes in Radiotherapy Regimens Incorporating Temozolomide or Gemcitabine in Glioblastoma. PLoS One 2012;7:e35857 [Crossref] [PubMed]
- Lemasson B, Wang H, Galbán S, et al. Evaluation of Concurrent Radiation, Temozolomide and ABT-888 Treatment Followed by Maintenance Therapy with Temozolomide and ABT-888 in a Genetically Engineered Glioblastoma Mouse Model. Neoplasia 2016;18:82-9. [Crossref] [PubMed]
- Halliday J, Helmy K, Pattwell SS, et al. In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural-mesenchymal shift. Proc Natl Acad Sci U S A 2014;111:5248-53. [Crossref] [PubMed]
- Hambardzumyan D, Becher OJ, Rosenblum MK, et al. PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes Dev 2008;22:436-48. [Crossref] [PubMed]
- Squatrito M, Brennan CW, Helmy K, et al. Loss of ATM/Chk2/p53 pathway components accelerates tumor development and contributes to radiation resistance in gliomas. Cancer Cell 2010;18:619-29. [Crossref] [PubMed]
- Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet 2005;6:507-12. [Crossref] [PubMed]
- Jinek M, Chylinski K, Fonfara I, et al. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012;337:816-21. [Crossref] [PubMed]
- Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science 2013;339:823-6. [Crossref] [PubMed]
- Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819-23. [Crossref] [PubMed]
- Ran FA, Hsu PD, Wright J, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8:2281-308. [Crossref] [PubMed]
- Jasin M, Haber JE. The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair. DNA Repair 2016;44:6-16. [Crossref] [PubMed]
- Gaj T, Gersbach CA, Barbas CF 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 2013;31:397-405. [Crossref] [PubMed]
- Sánchez-Rivera FJ, Papagiannakopoulos T, Romero R, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 2014;516:428-31. [Crossref] [PubMed]
- Xue W, Chen S, Yin H, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 2014;514:380-4. [Crossref] [PubMed]
- Maddalo D, Manchado E, Concepcion CP, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 2014;516:423-7. [Crossref] [PubMed]
- Platt RJ, Chen S, Zhou Y, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014;159:440-55. [Crossref] [PubMed]
- Blasco RB, Karaca E, Ambrogio C, et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep 2014;9:1219-27. [Crossref] [PubMed]
- Annunziato S, Kas SM, Nethe M, et al. Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes Dev 2016;30:1470-80. [Crossref] [PubMed]
- Maresch R, Mueller S, Veltkamp C, et al. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat Commun 2016;7:10770. [Crossref] [PubMed]
- Chiou SH, Winters IP, Wang J, et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev 2015;29:1576-85. [Crossref] [PubMed]
- Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun 2015;6:7391. [Crossref] [PubMed]
- Heckl D, Kowalczyk MS, Yudovich D, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol 2014;32:941-6. [Crossref] [PubMed]
- Kleinstiver BP, Prew MS, Tsai SQ, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015;523:481-5. [Crossref] [PubMed]
- Hsu PD, Scott DA, Weinstein JA, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013;31:827-32. [Crossref] [PubMed]
- Wu X, Scott DA, Kriz AJ, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 2014;32:670-6. [Crossref] [PubMed]
- Tycko J, Myer VE, Hsu PD. Methods for Optimizing CRISPR-Cas9 Genome Editing Specificity. Mol Cell 2016;63:355-70. [Crossref] [PubMed]
- Shen B, Zhang W, Zhang J, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods 2014;11:399-402. [Crossref] [PubMed]
- Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016;529:490-5. [Crossref] [PubMed]
- Slaymaker IM, Gao L, Zetsche B, et al. Rationally engineered Cas9 nucleases with improved specificity. Science 2016;351:84-8. [Crossref] [PubMed]
- Kim D, Bae S, Park J, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods 2015;12:237-43, 1 p following 243.
- Ran FA, Cong L, Yan WX, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015;520:186-91. [Crossref] [PubMed]
- Tsai SQ, Zheng Z, Nguyen NT, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 2015;33:187-97. [Crossref] [PubMed]
- Hilton IB, D’Ippolito AM, Vockley CM, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 2015;33:510-7. [Crossref] [PubMed]
- Thakore PI, D’Ippolito AM, Song L, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods 2015;12:1143-9. [Crossref] [PubMed]
- Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016;533:420-4. [Crossref] [PubMed]
- Chavez A, Scheiman J, Vora S, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods 2015;12:326-8. [Crossref] [PubMed]
- Chavez A, Tuttle M, Pruitt BW, et al. Comparison of Cas9 activators in multiple species. Nat Methods 2016;13:563-7. [Crossref] [PubMed]
- Gilbert LA, Larson MH, Morsut L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013;154:442-51. [Crossref] [PubMed]
- Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015;163:759-71. [Crossref] [PubMed]
- Abudayyeh OO, Gootenberg JS, Konermann S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016;353:aaf5573 [Crossref] [PubMed]
- East-Seletsky A, O’Connell MR, Knight SC, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016;538:270-3. [Crossref] [PubMed]
- Floyd SR, Pacold ME, Huang Q, et al. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature 2013;498:246-50. [Crossref] [PubMed]
- Adamson B, Smogorzewska A, Sigoillot FD, et al. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol 2012;14:318-28. [Crossref] [PubMed]
- Kondo S, Perrimon N. A genome-wide RNAi screen identifies core components of the G2-M DNA damage checkpoint. Sci Signal 2011;4:rs1. [Crossref] [PubMed]


