
What is the significance of TP53 and KRAS mutation for immunotherapy in non-small cell lung cancer?
Increasing evidences including PD-L1 expression (1,2), tumor mutation burden (TMB) (3,4), and the intensity of CD8+ T cell infiltrates (5-7) have been respectively certified as predictive biomarkers of response to immunotherapy. More recently, our study identified TP53/KRAS mutation may be another applicable factor to predict response to PD-1 blockade in non-small cell lung cancer (NSCLC) (8). This result has aroused some discussions. Dr. Tawee Tanvetyanon (9) raises a question of when is KRAS or TP53 mutation predictive of response to immunotherapy for lung cancer? He points out that it is not adequate enough to demonstrate the predictive value of KRAS mutation independently from TMB and that TP53 or KRAS mutation may be served as a predictive biomarker along with high TMB.
As the editorial mentioned, a number of genetic alternation, such as POLE, mismatch repair genes, BRCA1/2, and EGFR, strongly correlated with TMB, either positively or negatively, have been identified (10). In our study, we demonstrated tumor with TP53 and/or KRAS mutation showed significantly increased TMB and transversion mutation. We agree with that KRAS mutation cannot be separated from TMB in the prediction. We also haven’t enough data right now to verify the superiority of KRAS or TP53 mutation in those with a low TMB. One of the advantages in our study may be that TP53 or KRAS mutation could be routinely detected in the laboratory as a qualitative index. However, by comparison, it is relatively difficult to acquire the TMB and define a rational cut-off value. Even the PD-L1 protein detection still faced the perplexity of different antibody clones and cut-off values. Specific gene alteration might be available to substitute the TMB before which we can easily and precisely evaluate.
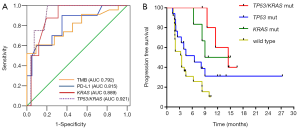
Apart from the convenient detection, the prediction of efficacy should be the leading consideration. Recent studies have explored the impact of co-mutations on the immune contexture and response of KRAS-mutant lung cancer to immunotherapy. Skoulidis et al. identified inactivation of STK11 is associated with establishment of “immune desert” in KRAS-mutant lung adenocarcinoma, while KRAS/TP53 co-mutation is associated with inflammation and active immunoediting (11). In their report, 35 KRAS-mutant NSCLC patients, consisting of 6 (17%) KRAS/STK11 and 17 (49%) KRAS/TP53 co-mutations, and 12 (34%) KRAS single mutation, underwent anti-PD-1 therapy. KRAS/TP53 group displayed a favorable objective response rate (ORR: 59%), followed by KRAS-single group (ORR: 25%), while KRAS/STK11 group without any response (ORR: 0). Meanwhile, KRAS/TP53 group also showed a prolonged progression free survival (PFS) than the other two groups (median: 23 vs. 16 vs. 6 weeks; P=0.0003) (12). Coincidently, we have demonstrated those with co-occurring TP53/KRAS mutations showed extremely increased PD-L1 expression, high density of CD8+ lymphocytes, and high TMB. Through the analysis of Memorial Sloan Kettering Cancer Center (MSKCC) cohort, we identified TP53 and KRAS co-mutation area under roc curve (AUC: 0.921; P<0.001) represented the optimal predictive factor of PFS of anti-PD-1 therapy in NSCLC, followed by KRAS mutation (AUC: 0.889), PD-L1-positive (PD-L1 ≥50%, AUC: 0.815), and high TMB (AUC: 0.792) (Figure 1A). Furthermore, through the analysis of total 54 patients from two cohorts [MSKCC and Guangdong Lung Cancer Institute (GLCI)], patients with TP53 and KRAS co-mutation showed the favorable clinical benefit than that of KRAS/TP53 single mutation and wild type patients (median: 14.5 vs. 12 vs. 6.5 vs. 3.5 months; P=0.0034; Figure 1B). These above results highlight the potential therapeutic vulnerabilities of the subgroup with TP53 and KRAS co-mutation. Nevertheless, there is still a long way to reach a precise immunotherapy. It is perhaps that we can combine two or more valuable and accessible biomarkers to reach an optimal prediction of immunotherapy in the future.
Acknowledgments
Funding: This study was supported by the Guangdong Provincial Key Laboratory of Lung Cancer Translational Medicine (Grant No. 2012A061400006) and the Special Fund for Research in the Public Interest from the National Health and Family Planning Commission of the People’s Republic of China (Grant No. 201402031).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Runzhe Chen (Department of Thoracic/Head & Neck Medical Oncology, University of Texas MD Anderson Cancer Center, Houston, TX, USA; Department of Hematology and Oncology, Zhongda Hospital of Southeast University, Nanjing, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.06.25). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515:563-7. [Crossref] [PubMed]
- Shukuya T, Carbone DP. Predictive Markers for the Efficacy of Anti-PD-1/PD-L1 Antibodies in Lung Cancer. J Thorac Oncol 2016;11:976-88. [Crossref] [PubMed]
- Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189-99. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Tang H, Wang Y, Chlewicki LK, et al. Facilitating T Cell Infiltration in Tumor Microenvironment Overcomes Resistance to PD-L1 Blockade. Cancer Cell 2016;29:285-96. [Crossref] [PubMed]
- Dong ZY, Wu SP, Liao RQ, et al. Potential biomarker for checkpoint blockade immunotherapy and treatment strategy. Tumour Biol 2016;37:4251-61. [Crossref] [PubMed]
- Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568-71. [Crossref] [PubMed]
- Dong ZY, Zhong WZ, Zhang XC, et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin Cancer Res 2017;23:3012-24. [Crossref] [PubMed]
- Tanvetyanon T. When is KRAS or TP53 mutation predictive of response to immunotherapy for lung cancer? Transl Cancer Res 2017;6:S424-6. [Crossref]
- Schrock A, Sharma N, Peled N, et al. Updated Dataset Assessing Tumor Mutation Burden (TMB) as a Biomarker for Response to PD-1/PD-L1 Targeted Therapies in Lung Cancer (LC). J Thorac Oncol 2017;12:S422. [Crossref]
- Skoulidis F, Byers LA, Diao L, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015;5:860-77. [Crossref] [PubMed]
- Skoulidis F, Elamin Y, Papadimitrakopoulou V, et al. Impact of major co-mutations on the immune contexture and response of KRAS-mutant lung adenocarcinoma to immunotherapy. WCLC 2016;17:abstr 6343.


