
Circulating exosomes and their cargos in blood as novel biomarkers for cancer
Introduction
Early diagnosis and real-time monitoring of therapeutic response could facilitate timely and efficient management of cancer patients. In this respect, many efforts have been made in developing minimally invasive, blood-based informative assays of liquid biopsies, such as circulating tumor cells (CTCs), circulating disseminated cells (CDCs), and circulating cell-free nucleic acids (RNAs and DNAs) (1-3). A decade ago, mRNAs and miRNAs were firstly identified in exosomes from the serum of patients with glioblastoma (GBM) (4) and ovarian cancer (5), which promotes the usage of exosomes and their contents as biomarkers for cancer. Since then, accumulating investigations started to focus on this fast-evolving research field.
Exosomes are nano-sized extracellular vesicles with a diameter range of 30–100 nm. In 1981, they were initially described and confirmed as microvesicles released from neoplastic cells (6-8). During the initial step of exosome formation, early multivesicular endosomes (MVEs) produced from cell membrane bud inward to form small internal vesicles carrying genetic materials and proteins. Some of the late MVEs fuse with the cell membrane and are released as exosomes into extracellular space (Figure 1). The extracellular exosomes mediate cell to cell communication via discharging their cargos to recipient cells, and thus influence the physiological and pathological functions of recipient cells (9-11). The functional investigation of exosomes originated from the description of exosome containing major histocompatibility complex class II (MHCII) secreted by antigen-presenting cells, implying their potential role in immune responses (12,13). Despite, all cell types release exosomes either under normal or pathological conditions, whereby tumor cells are the most avid producers (14). The characterization of exosomes carrying specific information dependent on their cell origin, and the relative facility to collect them from various biological fluids including blood, urine, tumor effusions, amniotic fluid or saliva (14), have suggested their use as diagnostic markers in pathology, in particular in cancer. Tumor-derived exosomes in blood are thought to promote tumor cell proliferation, angiogenesis and metastases by inhibiting antitumor immune responses, and thus, the depletion of these immune suppressive exosomes in blood circulation may be promising for the antitumor treatment (15,16). In addition, the cargos incorporated in exosomes are protected from degradation, which allow their quantification by various techniques. Thus, analyses of circulating exosomes and their contents in blood could be promising strategies to advance cancer diagnosis with better sensitivity and specificity, and monitor patients’ therapeutic response.
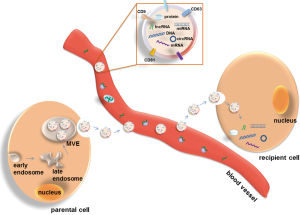
The property of exosomes in cancer
Considerable studies have suggested that tumor-derived exosomes influence multistep of tumor development, such as tumor initiation, growth, angiogenesis, hypoxia-driven epithelial-to-mesenchymal transition (EMT), metastasis, tumor immune escape, and drug resistance (17,18). For instance, GBM-derived exosomes which are enriched in angiogenic proteins have a capability to stimulate glioma cell proliferation as well as tubule formation (4). Exosomes derived from highly metastatic melanomas have been reported to increase the metastatic behavior of primary tumors by permanently “educating” bone marrow progenitors through the receptor tyrosine kinase MET. These melanoma-derived exosomes also induced vascular leakiness at pre-metastatic sites and reprogrammed bone marrow progenitors toward a pro-vasculogenic phenotype (19). Another direct evidence verifying the impact of tumor cell-derived exosomes on tumor progression was provided by Dutta and coworkers. They isolated exosomes from cholangiocarcinoma (CCA) cells, namely KKU-M213 and KKU-100, and incubated them with normal human cholangiocyte (H69) cells. They found that these cancer cell-derived exosomes were internalized into H69 cells and induced migration and invasion of H69 cells (20). Consistently, BRCA1-knockout human fibroblasts could be transformed into malignant cells after 2-week exposure to exosomes extracted from the sera of patients with breast, colorectal, pancreatic, and other cancer types (21). Recently, Nabet et al. reported that upon transfer to breast cancer cells, an endogenous RNA, unshielded RN7SL1 within stromal exosomes enhance tumor growth, metastasis and therapy resistance (22).
Therefore, these critical roles of tumor-derived exosomes in almost all aspects of cancer process provide opportunities for the development of exosome assays, and their functional cargos carrying tumor-specific signatures may be potential biomarker candidates in translational medicine. In this review, we refer the biofunctional cargos including proteins, miRNAs, lncRNAs, circRNAs, mRNAs and DNAs in exosomes to exo-proteins, exo-miRNAs, exo-lncRNAs, exo-circRNAs, exo-mRNAs and exo-DNAs, respectively.
Clinical relevance of circulating exosomes in cancer
Exosomes can be shed from various parental cells into blood, of which tumor cells are the most active producers. With concentrations as high as or higher than 109 vesicles/mL of blood, the number of exosomes secreted by tumor cells correlates with their malignant behavior. The relationship between circulating exosomes and tumor can be described as “seeds and plants”. Exosomes released by the parental cancer cells carrying multiple genetic materials are discharged into bloodstream. Some of circulating exosomes disseminate to distant organs and transform wild type cells to malignant ones, through transferring their bioactive contents into recipient cells (Figure 1). Thus, circulating exosomes may serve as a tool to detect tumor and predict treatment outcome, or as a therapeutic target.
In recent years, the clinical significance of circulating exosomes has been intensively investigated in blood of cancer patients. Tavoosidana and colleagues developed an extremely sensitive and specific assay, the modified proximity ligation assay (PLA) to detect exosomes in plasma of prostate cancer patients. They successfully detected significantly increased prostasomes (prostate specific exosomes) in patients with prostate cancer before radical prostatectomy, compared with controls and patients with benign prostate diseases, and especially, with a capability to distinguish high/medium Gleason scores from low ones (23). Similarly, Turay et al. quantified exosomes in plasma of prostate cancer patients using the acetylcholinesterase enzymatic assay and found exosome levels significantly higher in the prostate cancer cohort than the healthy men (24). Excessive active secretion of exosomes detected by CD63-based ELISA was observed in the serum of ovarian cancer patients compared to healthy women and patients with benign ovarian diseases (25). Melo and colleagues demonstrated that the relative concentrations of circulating exosomes were significantly higher in the sera of patients with breast cancer or pancreatic ductal adenocarcinoma compared to healthy individuals (26). Using nanoparticle tracking analysis (NTA), Arbelaiz et al. found higher concentrations of serum extracellular vesicles (mainly exosomes) in the hepatocellular carcinoma (HCC) cohort than healthy, intrahepatic CCA and primary sclerosing cholangitis (PSC) cohorts (27). By immunocapture-based microfluidic chip analysis, Fang and colleagues detected more epithelial cell adhesion molecule (EpCAM)-positive exosomes in patients with breast cancer (28).
Apart from the increase of circulating exosomes in the blood circulation of cancer patients, amazingly, the size and morphology alterations of exosomes can be also found in cancer. For instance, exosome size distribution analysis by NanoSight® showed that the size of serum exosomes derived from pancreatic ductal adenocarcinoma was significantly smaller than those from healthy donors (26). Similarly, using atomic force microscopy, Zlotogorski-Hurvitz et al. observed that oral fluid-derived exosomes from oral cancer patients differ both morphologically and molecularly from exosomes present in healthy donors (29). However, the underling mechanism of the morphological alterations of exosomes during tumor course is nubilous. Collectively, these investigations indicate that the amounts and morphological changes of exosomes in bloodstream are related to a specific course of the diseases, and may be used to estimate specific cancer status.
Compared to the detection of whole exosomes shed by all cell types, including malignant and wild type cells, into the blood circulation, it is more attractive and meaningful to detect tumor-derived exosomes, since they may reflect pathological alterations of parental malignancy, and thus, enable the specific identification of their contents without the contamination of non-cancer exosomes. However, the isolation of tumor-specific exosomes from the blood of cancer patients remains a challenge due to the lack of specific markers that can be used to identify and distinguish cancer exosomes from non-cancer-derived ones.
Clinical relevance of circulating exosomal cargos in cancer
Circulating exosomal proteins (exo-proteins)
Considering their endosomal origin, exosomes released from different cell types are enriched of endosome-associated proteins, like Rab GTPase, annexins and flotillin (30). Tetraspanins, such as CD63, CD81, CD82 and CD9, are components of the exosomal membrane and frequently used as exosome markers. ExoCarta, an exosome database (http://exocarta.org/), has been developed to identify the exosomal contents. Up to now, approximately 10,000 different proteins have been characterized (31,32). The protein composition in total exosome fractions isolated from blood is significantly higher in cancer patients than in healthy controls. For example, the protein concentration in serum-derived exosomes from patients with acute myeloid leukemia (AML) showed a value of around 75 µg protein/mL, and was 60-fold higher than those isolated from the serum of normal controls (about 1.2 µg protein/mL) (33).
Recently, studies uncovered that, based on the property of proteins, either as a tumor promoter or suppressor, the export of proteins by exosomes impacts tumor environment and is also linked to chemotherapy response. In patients with AML undergoing chemotherapy, the protein levels in exosomes derived from plasma went down on day 14 after the initiation of chemotherapy, whereas during consolidation therapy with high-dose cytarabine, the exo-protein levels increased to a similar level at diagnosis, indicating that the dynamic changes of exo-protein levels after induction and during consolidation chemotherapy could be useful for evaluating patients’ responses to chemotherapy (34). The study by Lv’s group showed that anticancer drugs enhanced the release of heat shock proteins (HSPs)-enriched exosomes from HCC cells that elicited effective natural killer cell antitumor responses in vitro (35). Previously, Safaei and coworkers revealed that cisplatin-resistant ovarian cancer cells released exosomes that contained an excess of proteins and also carried the cisplatin export transporters, MRP2 (mitochondrial 37S ribosomal protein), ATP7A (ATPase copper transporting alpha) and ATP7B (ATPase copper transporting beta). Following the exposure to cisplatin, the exosomes released from cisplatin-resistant cells contained 2.6-fold more platinum than those released from sensitive cells, indicating the active export of chemotherapy drug by exosomes (36). Consistently, the study by Szajnik et al. observed that patients with ovarian cancer whose exo-protein levels decreased after therapy were clinical responders to chemotherapy, suggesting that exo-proteins in blood could act as a potential biomarker to predict response to chemotherapy (37). In addition, exosomes released by the HER2-overexpressing breast cancer cell lines could discharge a full-length active HER2 molecule, resulting in resistance to Trastuzumab and implying the association of HER2-positive exosomes and sensitivity to Trastuzumab in breast cancer (38).
Besides their mediation of therapeutic response to anti-cancer drugs, exo-proteins also contribute to other actions during tumor development. Putz et al. showed that phosphatase and tensin homolog (PTEN), an important tumor suppressor protein, could be taken up via exosomes by recipient cells with resultant functional phosphatase activity, leading to the repression of cell proliferation in GBM (39). Annexin II, which was highly expressed in exosomes from malignant and premetastatic breast cancer cells was proved to promote tissue plasminogen activator (tPA)-dependent angiogenesis and facilitate the formation of favorable microenvironment for metastasis, via activating macrophage-mediated p38MAPK (p38 map kinase), NF-κB (nuclear factor kappa B), and STAT3 (signal transducer and activator of transcription 3) pathways and increasing secretion of IL6 (interleukin 6) and TNFα (tumor necrosis factor alpha) (40). While studying the role of Wnt5b in cancer progression, Harada and colleagues found that 55% of secreted endogenous Wnt5b was associated with exosomes. These exosomes containing activated Wnt5b proteins stimulated migration and proliferation of A549 lung adenocarcinoma cells (41). In summary, these findings open new and interesting avenues for the investigation of tumor-associated proteins not only at original tumor sites but also in distant metastatic organs where these proteins are trafficked by exosomes in a paracrine manner.
Concerning the vital roles of proteins within exosomes, proteomic analyses of whole exosomes, in particular tumor-derived exosomes have been performed to screen biomarkers for cancer (Table 1). For instance, by quantitative proteomics analysis, Chen et al. identified 36 up-regulated proteins involved in processes that modulate the protumorigenic microenvironment for metastasis, and 22 down-regulated proteins related to tumor growth and cell survival, in the serum-purified exosomes from patients with colorectal cancer. These findings suggest the utility of deregulated exo-proteins in the blood-based diagnosis of cancer (49). An and coworkers analyzed the exo-proteome in the serum of patients with pancreatic cancer undergoing chemoradiotherapy at different time points and identified 8 proteins that exhibited global treatment-specific changes, implying their potential to monitor treatment resistance during the course of therapy (43). Database-augmented mass spectrometry analysis of exosomes isolated from plasma of patients with prostate cancer conducted by Worst et al. revealed that the amounts of the tight junction protein claudin 3 were higher in patients with Gleason ≥8 tumors than patients with benign prostatic hyperplasia (BPH) and Gleason 6–7 tumors. The levels of this exo-protein could predict a Gleason score ≥8 with an area under the curve (AUC) value of 0.705, suggesting the diagnostic power of exo-claudin 3 in prostate cancer (53). Arbelaiz et al. determined the protein composition of serum exosomes isolated from CCA, PSC, HCC and healthy individuals by mass spectrometry. In this profiling study, the researchers screened 95 exo-proteins differentially expressed in CCA vs. controls, 161 in PSC vs. controls, 50 in CCA vs. PSC, and 98 in HCC vs. controls, which are mostly related to response to wounding, defense to infections, inflammatory responses and immune activation. Specifically, among these differentially expressed exo-proteins, aminopeptidase N (AMPN), pantetheinase (VNN1) and polymeric immunoglobulin receptor (PIGR) showed the best diagnostic capacity to differ CCA and control with AUC values of 0.878, 0.876 and 0.844, respectively. Notably, galectin-3-binding protein (LG3BP) and PIGR present better diagnostic capability than alpha-fetoprotein (AFP), a non-specific serum tumor biomarker commonly employed in HCC, to discriminate HCC from healthy individuals (27). This study indicates that these multiple differentially expressed oncogenic proteins confined in exosomes can be utilized as diagnostic tools in cancer. Kawakami and coworkers observed higher gamma-glutamyltransferase activity in exosomes derived from the serum of patients with prostate cancer than patients with BPH (54). In the serum of patients with AML, exosome-associated transforming growth factor-β1 (TGF-β1) was significantly higher than that in normal controls (33). Following this discovery, Hong et al. firstly revealed the potential link between TGF-β1 levels in plasma-derived exosomes and therapeutic response in patients with AML. Exo-TGF-β1 levels at AML diagnosis were much higher than those of healthy donors, and then reduced drastically following induction of chemotherapy, but subsequently found to be elevated relative to post-induction levels in patients receiving consolidation (34). Furthermore, the researchers also observed 3 different forms of TGF-β1 [the pro-peptide, latency-associated peptide (LAP) or active, mature form] in plasma exosomes from patients with AML before, during, or after chemotherapy. For instance, apart from the distinctly different levels of active TGF-β1 in exosomes obtained before, during or after chemotherapy, the levels of exo-LAP expression were generally high at AML diagnosis and also in exosomes of patients undergoing consolidation chemotherapy, indicating that exo-TGF-β1 levels and TGF-β1 activation in exosomes might reflect response to chemotherapy (34). The same lab also found elevated total protein levels in plasma exosomes of patients with malignant glioma, and their positive correlation with WHO tumor grade (57). Moreover, exo-protein levels were measured in the plasma of patients who received phase I/II dendritic cell/peptide-based vaccination treatment. The protein levels exhibited a rapid decrease from pre- to post-vaccination, and were related to immunological and clinical response, implying exo-proteins as a predictive marker of clinical response to vaccine (57). In patients with prostate cancer, the total protein levels contained in exosomes isolated from serum were abnormally increased in docetaxel resistant patient subset compared to the sensitive group. Specifically, western blot analysis showed that MDR-1/3 (multidrug transporter-1/3) and PABP4 (poly A binding protein 4) were exclusively overexpressed in the patients with docetaxel resistant prostate cancer, indicating their value to predict response to chemotherapy (55). Moreover, exosomes from the serum of patients with metastatic melanoma were observed to contain higher levels of MDA-9 and GRP78 proteins than those of patients without metastases, suggesting their utilities as biomarkers for early detection of metastasis (46). PTEN is frequently reduced or completely lost in aggressive and metastatic prostate cancer. However, PTEN has been detected in plasma-derived exosomes from patients with prostate cancer but not healthy subjects, suggesting that exo-PTEN may compensate for PTEN loss in advanced prostate cancer, and may have diagnostic value for prostate cancer (51). Khan and colleagues showed much higher levels of exo-Survivin in plasma of patients with prostate cancer, especially in relapsed patients on chemotherapy than patients with BPH and healthy individuals (52), indicating the association of exo-Survivin and sensitivity to chemotherapeutic agents in prostate cancer. Using anti-CD9 antibody-coupled highly porous monolithic silica microtips, Ueda et al. successfully captured exosomes from the serum of 46 patients with lung cancer. The subsequent mass spectrometric analysis identified CD91 as a lung adenocarcinoma specific antigen on exosomes to distinguish lung cancer from healthy individuals (58). In melanoma, the levels of 4 proteins, tyrosinase-related protein 2 (TYRP2), very late antigen-4 (VLA-4), heat shock protein 70 (HSP70) and MET oncoprotein were increased in plasma exosomes from stage III and IV patients compared to stage I patients as well as healthy controls (19). Moreover, the high protein content of plasma-derived exosomes correlated with poor survival (19). Prostate specific antigen (PSA) is the most important tumor marker in prostate cancer. Logozzi et al. detected that, compared to benign prostatic hypertrophy, patients with prostate cancer had high levels of exosomes expressing both CD81 and PSA in plasma, implying that PSA-exosomes, may represent a novel clinical tool for the screening of prostate cancer (56).

Full table
To sum up, the identification of specific exo-proteins can not only facilitate diagnosis, prognosis and monitoring chemotherapeutic response, but also contribute to the screening of specific antigens characteristically expressed on tumor-derived exosomes, which can be used as makers to capture tumor specific exosomes.
Circulating exosomal miRNAs (exo-miRNAs)
miRNAs, the small non-coding RNAs of 18–25 nucleotides in length, are involved in the course of tumor development and progression via binding specifically to the 3' UTR of their target mRNAs, leading to posttranscriptional repression of tumor-associated mRNAs (59). MiRNAs can be released into bloodstream as molecules incorporated in exosomes, or as cell-free miRNAs bound to argonaute (AGO) proteins or high-density lipoproteins (HDL) (2), which prevent them from the digestion of RNase and thus, exist stably in blood circulation. Owing to their important roles in cancer and their stability in blood, the utility of miRNAs as “liquid biopsy” for cancer detection has been intensively investigated. In bloodstream, miRNAs integrated in exosomes account for the largest proportion of circulating miRNAs discharged from different sources (60). Therefore, it is of great interest to investigate the patterns of circulating exo-miRNAs in cancer patients.
For example, Taylor et al. demonstrated that miR-21, miR-203, miR-205, miR-214 and miR-200 family (with the exception of miR-429) were selectively sorted into EpCAM-specific exosomes in the serum of ovarian cancer patients (5). In patients with breast cancer, serum exo-miR-373 seemed to have the capability to distinguish different molecular subtypes. Specifically, its levels were much higher in triple negative than luminal carcinomas. Also, estrogen and progesterone-negative patients harbored more serum exo-miR-373 than those with hormone-receptor positive tumors. These data suggest that serum exo-miR-373 correlates with receptor negative subtype and may contribute to the progression of breast cancer (61). Another study with a cohort of 163 patients with epithelial ovarian cancer (EOC) was conducted in the same lab. The patients with EOC displayed increased serum levels of exo-miR-373, which further verifies the prevalence of this oncogenic miRNAs in different cancers (25). Moreover, the study also showed that serum levels of exo-miR-200a, miR-200b and miR-200c differ between benign and malignant ovarian tumors, and the combination of the three exo-miRNAs could improve the sensitivity to discriminate benign form malignant ovarian tumors (25).
Apart from breast cancer (61) and ovarian cancer (5,25), previous studies have reported that circulating exo-miRNAs in blood of various cancer types, including lung (62-66), thyroid (67), esophageal adenocarcinoma (68), esophagus adenocarcinoma (69,70), colon (71-77), hepatoblastoma (78), hepatocellular (79,80), pancreatic (81,82), prostate (83-85), multiple myeloma (86), leukemia (87,88), myeloma (89), melanoma (90), laryngeal squamous cell carcinoma (LSCC) (91) and osteosarcoma (92), also show distinct signatures, which may be used for cancer diagnosis, prognosis and monitoring response to chemotherapy (Table 2).
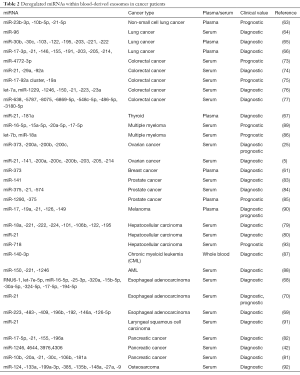
Full table
The above exo-miRNA profiling showed that during tumor procession, a subset of miRNAs was preferentially incorporated into exosomes, indicating a selective miRNA export mechanism by exosomes (94). However, whether the miRNA transport is rather selective or non-selective, is still arguable, and needs to be further investigated.
Still, there is no consensus on an endogenous control for the normalization of exo-miRNAs as well as cell-free miRNAs, which may lead to discrepant results among different studies, or even inaccurate results. For an accurate analysis of circulating miRNAs, a comprehensive screening of the relatively consistent miRNAs that can be used as endogenous normalizers is highly recommended. Moreover, the combination of endogenous and exogenous spike-in controls can minimalize the bias caused by the quality of isolated miRNAs, and the sample preparations and experimental handling (95).
Circulating exosomal long non-coding RNAs (exo-lncRNAs)
Long non-coding RNAs are RNA molecules more than 200 nucleotides in length. They have been reported to have diverse functions in the regulation of gene expression, e.g., epigenetic regulation, chromatin modification, transcription or posttranscriptional processing (96). It is becoming increasingly clear lncRNAs are crucial regulators of processes involving the initiation and progression of tumors. For example, the lncRNA-activated by TGF-β (lncRNA-ATB) up-regulated ZEB1 and 2 (zinc finger E-box binding homeobox 1 and 2) through competitively binding with miR-200 family members to induce EMT and cell invasion. Moreover, lncRNA-ATB promoted organ colonization of disseminated tumor cells (DTCs) by binding with IL-11mRNA to trigger STAT3 signaling (97). Ahadi and colleagues examined lncRNAs and miRNAs enriched in exosomes that were released into the cell culture media by prostate cancer cell lines. They uncovered that 26 exo-lncRNAs are enriched for miRNA seeds with a preference for let-7 family members as well as miR-17, miR-18a, miR-20a, miR-93 and miR-106b, indicating the potential of exo-lncRNAs as miRNA sponges in tumor-related gene regulation (98). Accumulating evidence is revealing an important role of lncRNAs in tumor drug resistance. For instance, Wang’s research group initially identified and named lncARSR (lncRNA activated in renal cell carcinoma with sunitinib resistance), which promoted sunitinib resistance via competitively binding with miR-34/miR-449 to facilitate the expressions of receptor tyrosine kinase AXL and c-MET in renal cancer cells. Intriguingly, the bioactive lncARSR could be incorporated into exosomes and transmitted to sensitive cells to transfer sunitinib resistance (99).
Currently, only a few reports regarding the utility of circulating exo-lncRNAs as biomarkers for cancer are available (Table 3). Dong and coworkers reported the presence of 21 cancer-related lncRNAs differentially expressed in serum exosomes of patients with colorectal cancer compared to healthy controls, indicating the potential of exo-lncRNAs to diagnose colorectal cancer (102). MALAT-1 (metastasis-associated lung adenocarcinoma transcript) is the first identified lncRNA that is related to lung cancer metastasis. Zhang et al. measured significantly higher levels of MALAT-1 in serum exosomes of patients with non-small cell lung cancer (NSCLC) than healthy donors. More importantly, the elevated levels of exo-MALAT-1 were positively associated with tumor stage and lymphatic metastasis (104). As a newly identified lncRNA, lncZFAS1 (zinc finger antisense 1) promoted cancer cell proliferation and migration by activating the expressions of ZEB1, MMP14 and MMP16 (matrix metalloproteinase 14 and 16) in HCC (106). In the serum of patients with gastric cancer, the increased levels of exo-lncZFAS1 significantly correlated with lymphatic metastasis and TNM stage (100). In bloodstream of patients with gastric cancer, the lncRNA, LINC00152 was reported to be mainly incorporated in exosomes. The levels of exo-LINC00152 were significantly elevated in patients with gastric cancer compared with healthy controls (101). In the serum of patients with colorectal cancer, Liu at al. found that exo-CRNDE-h levels significantly correlated with regional lymph node metastasis and distant metastasis. In addition, high exo-CRNDE-h levels were associated with a shorter patients’ overall survival (103). In the serum of LSCC, the amounts of exo-miR-21 and exo-lncRNA HOTAIR were significantly higher in patients with LSCC than those with vocal cord polyps. In addition, higher serum exo-miR-21 and exo-HOTAIR expressions were linked to tumor stages and status of lymph node metastasis. Interestingly, the combination of exo-miR-21 and exo-HOTAIR could improve the sensitivity and specificity to detect LSCC (91). To sum up, these tumor-associated exo-lncRNAs are promising cancer biomarkers.
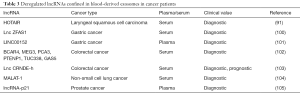
Full table
Circulating exosomal circular RNAs (exo-circRNAs)
Circular RNAs (circRNAs) are a large class of non-coding RNAs that, unlike linear RNAs, form covalently closed continuous loops. They participate in cancer process, mainly through acting as miRNA sponges to indirectly influence miRNA targets, enhancing the expression of their parental mRNAs at transcriptional level and sequestering RNA-binding proteins or ribonucleoprotein complexes related to tumorigenesis and progression (107-110). The expression levels of circRNAs have been found to be deregulated in all types of cancer cell lines, tumor tissues, as well as blood samples from patients, and linked to certain clinical characteristics (111,112), suggesting their involvement in tumor progression.
The presence of abundant circRNAs within exosomes was firstly reported by Li and colleagues (113). They reported that circRNAs are more abundant in exosomes compared to the cytoplasm of producer cells. Taking advantage of RNA-seq analyses and qRT-PCR, this research group identified 1,215 circRNAs in human serum, and pointed out that, these circulating circRNAs in bloodstream were predominantly located in serum-derived exosomes but not in exosome-depleted serum. Furthermore, they observed that abundance of tumor-derived exo-circRNAs in the serum of patients with colorectal cancer was correlated with tumor mass. They also found that the expression profile of exo-circRNAs in cancer serum was significantly different from that in normal serum. Specifically, the expression levels of circ-KLDHC10 were significantly increased in cancer serum than those in healthy individuals, indicating its diagnostic potential. Furthermore, concerning the sorting of specific circRNAs in exosomes, Li and colleague provided evidence that sorting of circRNAs in exosomes may be regulated by changes of associated miRNA levels in producer cells (113). Consistently, Zhang’s research group also observed more abundant circRNAs in exosomes than in cells. Besides, they detected significantly down-regulated levels of circRNAs in KRAS mutant colon cancer cells compared to wild-type cells, indicating that circRNAs enclosed in exosomes may be associated with KRAS status in colon cancer (114). Apart from these both investigations, so far, no further reports concerning exo-circRNAs are available. However, the research interest is now shifting from miRNAs and lncRNAs to circRNAs.
Circulating exosomal mRNAs (exo-mRNAs)
The presence of mRNAs in exosomes and their horizontal transfer via exosome vehicle to recipient cells were firstly elucidated by Valadi and coworkers in 2007 (10). Bioactive mRNAs (oncogenes or tumor suppressor genes) incorporated in exosomes were demonstrated to be delivered to target cells and translated into functional proteins, to change the phenotype of targeted cells, and even, further transform normal to malignant cells (4,10,115). Consequently, the abnormal expression of these tumor-associated exo-mRNAs encapsulated in exosomes present a potential to be used as predictive markers in cancer (Table 4).
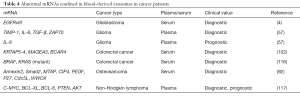
Full table
For instance, Skog et al. reported that serum exosomes from patients with GBM contained over 4,700 distinct mRNAs, of which tumor-specific EGFRvIII RNA was detected in 7 out of 25 patients with GBM (4). Selected RNAs, TIMP-1 (tissue inhibitor of metalloproteinase 1), IL-8, TGF-β and ZAP70 (zeta chain of T-cell receptor associated protein 70), which are related to angiogenesis and clinical outcome in gliomas were quantified in exosomes isolated from plasma of patients with glioma at baseline and 8 weeks after vaccination therapy by the Whiteside’s group. The four mRNAs were highly expressed at baseline and strongly down-regulated after vaccination (57). Recently, Hao and colleagues detected three mutations in the KRAS (Kirsten rat sarcoma viral oncogene) mRNA (codons 12, 13 and 61) and a mutation in BRAF (B-Raf proto-oncogene, serine/threonine kinase) mRNA (codon 600) in the serum exosomes of patients with colorectal cancer of a Chinese population (116). Dong et al. quantified the expression levels of 3 exo-mRNAs, keratin associated protein 5-4 (KRTAP5-4), melanoma antigen family A3 (MAGEA3) and breast cancer anti-estrogen resistance 4 (BCAR4), confined in the serum exosomes of patients with colorectal cancer. They found that the combination of the three exo-mRNAs were able to detect colorectal cancer with an AUC of 0.936 (102). While profiling the miRNA signature in serum exosomes from patients with osteosarcoma, Xu and coworkers observed that Annexin2, Smad2, methylthioadenosine phosphorylase (MTAP), Cdc42-interacting protein 4 (CIP4), pigment epithelium-derived factor (PEDF), WW domain-containing oxidoreductase (WWOX), cell division cycle 5-like (Cdc5L) and p27 mRNAs were differentially expressed in exosomes, and associated with a different chemotherapeutic response (92). Additionally, a recent report demonstrated the diagnostic and prognostic relevance of exo-mRNAs of c-MYC (c-MYC proto-oncogene, bHLH transcription factor), BCL-XL, BCL-6, PTEN and AKT (AKT serine/threonine kinase) in the plasma of patients with non-Hodgkin lymphoma (117). Specifically, the presence of AKT mRNA was associated with a poor response to rituximab-based treatment. The lower levels of PTEN and BCL-XL mRNAs were associated with relapse or disease progression. The presence of c-MYC mRNA correlated with a short progression-free survival. BCL-6 and c-MYC mRNAs were independent prognostic variables of overall survival.
Despite their less abundance than miRNAs, the tumor-related exo-mRNAs carry important information on expression patterns and presence of mutations and variants, indicating that they can be developed as surrogates of cancer diagnosis and targeted therapies.
Circulating exosomal DNAs (exo-DNAs)
Besides miRNAs, lncRNAs, circRNAs and mRNAs within exosomes, distinct types of DNAs (single-stranded DNAs, double-stranded DNAs and mitochondrial DNAs) are incorporated in exosomes, of which double-stranded DNAs account for the majority of DNA types (118). Genomic DNA (gDNA) fragments transferred from exosomes to cells have been shown not only to increase the gDNA-coding mRNA and protein levels, but also to influence functions in recipient cells (119). It has been demonstrated by Thakur et al. that exo-DNAs represent the entire genome and reflect the mutational status of parental tumor cells (120). Thus, the investigation of molecular alterations of tumor-related exo-DNAs is an attractive strategy in the early detection of cancers and monitoring of treatment response (Table 5).

Full table
Using PCR-based DNA fragment analysis, Fricke et al. revealed frameshift mutation patterns in microsatellite stretches of microsatellite unstable (MSI) target genes, MARCKS (myristoylated alanine rich protein kinase C substrate), TGFBR2 (transforming growth factor beta receptor 2) and LMAN1(lectin, mannose binding 1) in exosomes released by colorectal cancer cell lines harboring MSI, indicating that the coding MSI phenotype of DNAs is maintained in the exo-DNAs (124). By whole genome sequencing, Kahlert et al. demonstrated that serum exosomes from patients with pancreatic cancer contain gDNA spanning all chromosomes with mutated KRAS and p53, indicating that serum-derived exosomes can be used to determine gDNA mutations for cancer detection, treatment, and therapy resistance (121). Similarly, Yang and colleagues also detected the presence of mutant KRAS and p53 in serum exosomes of patients with pancreatic cancer (122). Consistently, another study also reported a highly frequent mutation of KRAS in exo-DNAs in the plasma of patients with pancreatic cancer. These higher mutation allele frequencies of exo-KRAS DNA were associated with decreased disease-free survival in the patients with localized pancreatic cancer (123).
In summary, exo-DNAs in cancer may indeed represent a new treasure trove for monitoring changes in the mutational landscape to predict cancer presence, prognosis or response to chemotherapy. However, the issues associated with the diagnostic sensitivity and noise signal due to the contamination of tumor-specific DNAs with DNAs from normal cells are a challenge. Therefore, it is important to selectively enrich tumor-specific exosomes prior to the determination of translational value of exo-DNAs as liquid biopsy.
Conclusions
As the knowledge of biogenesis, biomolecule cargos and biological functions of exosomes, in particular tumor cell-derived exosomes, advances, the documents about exploiting exosomes and their cargos as “liquid biopsy” in real-time, to monitor cancer development and progression, are accumulating. However, a challenging aspect with respect to the exploration of exosomes-based biomarkers has turned out to be their effective and selective isolation. The plurality of developed isolation protocols including ultracentrifugation, chemical precipitation, affinity capturing, filtration techniques, and ultracentrifugation combined with iodixanol density gradient centrifugation or gel filtration results in qualitative and quantitative variability in terms of the extracted exosomes, which may lead to discrepant results of downstream analyses and make it difficult to compare or reproduce the results among different research groups. Tumor-specific exosomes, as a subpopulation of whole exosomes released by tumor cells, are distinct from exosomes secreted by normal cells, and may, of course, provide more information of tumor-related pathways. The identification of tumor-specific markers on exosomes with functional properties reflecting the characteristics of the parental tumor cells has been pressed ahead. Although remarkable progress in bringing exosomes, particularly tumor-specific exosomes to the attention of the scientific and clinical communities has been made, assays of exosomes in large patient cohorts with an exact description of the clinical pathological parameters (tumor stage, metastatic status, response to chemotherapy, long-term follow up) need to be conducted under standard criteria. All these hurdles have to be overcome before the clinical application of tumor-specific exosomes as a “liquid biopsy” in cancer patients.
Besides their potential value as biomarker, and based on their unique characteristics as stable, small molecules to cross the blood-brain barrier, exosomes can be created as carriers for the delivery of small interfering RNA (siRNA) or drugs, which have been proved to be effective in pre-clinical studies and clinical trials. In this regard, dendritic cell-derived exosomes as immunotherapeutic anticancer agents have entered clinical trials for colorectal cancer, metastatic melanoma and NSCLC. However, they have only achieved modest therapeutic effects (125,126). Moreover, due to the oncogenic property of tumor-associated exosomes to transform normal to malignant cells, the depletion of exosomes from blood of advanced cancer patients is a novel strategy to treat cancer (127).
Since exosomes are small particles with big roles in cancer, they are promising biomarkers as well as target molecules for the clinical practice.
Acknowledgments
Funding: This work was supported by the Zhejiang Provincial Natural Science Foundation of China under Grant No. LQ18H200001, LY15C060003; the National Undergraduate Training Program for Innovation and Entrepreneurship (201711646018); the Scientific Innovation Team Project of Ningbo (No. 2017C110019); the Non-profit Technology Research Program of Zhejiang Province (LGF18H160006); the Natural Science Foundation of Ningbo (2017A610247); the Sci-Tech Research Project of Ningbo City (No. 2016C51019) and the K.C. Wong Magna Fund in Ningbo University.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Heidi Schwarzenbach) for the series “Technologies in Liquid Biopsies - Potential applications in Medicine” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.09.17). The series “Technologies in Liquid Biopsies - Potential applications in Medicine” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0.
References
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. [Crossref] [PubMed]
- Schwarzenbach H, Nishida N, Calin GA, et al. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol 2014;11:145-56. [Crossref] [PubMed]
- Quandt D, Dieter Zucht H, Amann A, et al. Implementing liquid biopsies into clinical decision making for cancer immunotherapy. Oncotarget 2017;8:48507-20. [Crossref] [PubMed]
- Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008;10:1470-6. [Crossref] [PubMed]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 2008;110:13-21. [Crossref] [PubMed]
- Trams EG, Lauter CJ, Salem N Jr, et al. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta 1981;645:63-70. [Crossref] [PubMed]
- Pan BT, Teng K, Wu C, et al. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol 1985;101:942-8. [Crossref] [PubMed]
- Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol 1983;97:329-39. [Crossref] [PubMed]
- Théry C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep 2011;3:15. [Crossref] [PubMed]
- Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Ratajczak J, Wysoczynski M, Hayek F, et al. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia 2006;20:1487-95. [Crossref] [PubMed]
- Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med 1996;183:1161-72. [Crossref] [PubMed]
- Zitvogel L, Regnault A, Lozier A, et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med 1998;4:594-600. [Crossref] [PubMed]
- Chaput N, Thery C. Exosomes: immune properties and potential clinical implementations. Semin Immunopathol 2011;33:419-40. [Crossref] [PubMed]
- Ichim TE, Zhong Z, Kaushal S, et al. Exosomes as a tumor immune escape mechanism: possible therapeutic implications. J Transl Med 2008;6:37. [Crossref] [PubMed]
- Fan GC. Hypoxic exosomes promote angiogenesis. Blood 2014;124:3669-70. [Crossref] [PubMed]
- Zhang X, Yuan X, Shi H, et al. Exosomes in cancer: small particle, big player. J Hematol Oncol 2015;8:83-95. [Crossref] [PubMed]
- Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev 2013;32:623-42. [Crossref] [PubMed]
- Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012;18:883-91. [Crossref] [PubMed]
- Dutta S, Reamtong O, Panvongsa W, et al. Proteomics profiling of cholangiocarcinoma exosomes: A potential role of oncogenic protein transferring in cancer progression. Biochim Biophys Acta 2015;1852:1989-99. [Crossref] [PubMed]
- Hamam D, Abdouh M, Gao ZH, et al. Transfer of malignant trait to BRCA1 deficient human fibroblasts following exposure to serum of cancer patients. J Exp Clin Cancer Res 2016;35:80-91. [Crossref] [PubMed]
- Nabet BY, Qiu Y, Shabason JE, et al. Exosome RNA Unshielding Couples Stromal Activation to Pattern Recognition Receptor Signaling in Cancer. Cell 2017;170:352-66.e13. [Crossref] [PubMed]
- Tavoosidana G, Ronquist G, Darmanis S, et al. Multiple recognition assay reveals prostasomes as promising plasma biomarkers for prostate cancer. Proc Natl Acad Sci U S A 2011;108:8809-14. [Crossref] [PubMed]
- Turay D, Khan S, Diaz Osterman CJ, et al. Proteomic Profiling of Serum-Derived Exosomes from Ethnically Diverse Prostate Cancer Patients. Cancer Invest 2016;34:1-11. [Crossref] [PubMed]
- Meng X, Mueller V, Milde-Langosch K, et al. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget 2016;7:16923-35. [Crossref] [PubMed]
- Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015;523:177-82. [Crossref] [PubMed]
- Arbelaiz A, Azkargorta M, Krawczyk M, et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology 2017;66:1125-43. [Crossref] [PubMed]
- Fang S, Tian H, Li X, et al. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PLoS One 2017;12:e0175050 [Crossref] [PubMed]
- Zlotogorski-Hurvitz A, Dayan D, Chaushu G, et al. Morphological and molecular features of oral fluid-derived exosomes: oral cancer patients versus healthy individuals. J Cancer Res Clin Oncol 2016;142:101-10. [Crossref] [PubMed]
- Raposo G, Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J Cell Biol 2013;200:373-83. [Crossref] [PubMed]
- Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 2009;9:4997-5000. [Crossref] [PubMed]
- Mathivanan S, Fahner CJ, Reid GE, et al. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res 2012;40:D1241-4. [Crossref] [PubMed]
- Szczepanski MJ, Szajnik M, Welsh A, et al. Blast-derived microvesicles in sera from patients with acute myeloid leukemia suppress natural killer cell function via membrane-associated transforming growth factor-beta1. Haematologica 2011;96:1302-9. [Crossref] [PubMed]
- Hong CS, Muller L, Whiteside TL, et al. Plasma exosomes as markers of therapeutic response in patients with acute myeloid leukemia. Front Immunol 2014;5:160-8. [Crossref] [PubMed]
- Lv LH, Wan YL, Lin Y, et al. Anticancer drugs cause release of exosomes with heat shock proteins from human hepatocellular carcinoma cells that elicit effective natural killer cell antitumor responses in vitro. J Biol Chem 2012;287:15874-85. [Crossref] [PubMed]
- Safaei R, Larson BJ, Cheng TC, et al. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther 2005;4:1595-604. [Crossref] [PubMed]
- Szajnik M, Derbis M, Lach M, et al. Exosomes in Plasma of Patients with Ovarian Carcinoma: Potential Biomarkers of Tumor Progression and Response to Therapy. Gynecol Obstet (Sunnyvale) 2013;3. [PubMed]
- Ciravolo V, Huber V, Ghedini GC, et al. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J Cell Physiol 2012;227:658-67. [Crossref] [PubMed]
- Putz U, Howitt J, Doan A, et al. The tumor suppressor PTEN is exported in exosomes and has phosphatase activity in recipient cells. Sci Signal 2012;5:ra70. [Crossref] [PubMed]
- Maji S, Chaudhary P, Akopova I, et al. Exosomal Annexin II Promotes Angiogenesis and Breast Cancer Metastasis. Mol Cancer Res 2017;15:93-105. [Crossref] [PubMed]
- Harada T, Yamamoto H, Kishida S, et al. Wnt5b-associated exosomes promote cancer cell migration and proliferation. Cancer Sci 2017;108:42-52. [Crossref] [PubMed]
- Madhavan B, Yue S, Galli U, et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer 2015;136:2616-27. [Crossref] [PubMed]
- An M, Lohse I, Tan Z, et al. Quantitative Proteomic Analysis of Serum Exosomes from Patients with Locally Advanced Pancreatic Cancer Undergoing Chemoradiotherapy. J Proteome Res 2017;16:1763-72. [Crossref] [PubMed]
- Klibi J, Niki T, Riedel A, et al. Blood diffusion and Th1-suppressive effects of galectin-9-containing exosomes released by Epstein-Barr virus-infected nasopharyngeal carcinoma cells. Blood 2009;113:1957-66. [Crossref] [PubMed]
- Logozzi M, De Milito A, Lugini L, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One 2009;4:e5219 [Crossref] [PubMed]
- Guan M, Chen X, Ma Y, et al. MDA-9 and GRP78 as potential diagnostic biomarkers for early detection of melanoma metastasis. Tumour Biol 2015;36:2973-82. [Crossref] [PubMed]
- Hong CS, Muller L, Boyiadzis M, et al. Isolation and characterization of CD34+ blast-derived exosomes in acute myeloid leukemia. PLoS One 2014;9:e103310 [Crossref] [PubMed]
- Yoshioka Y, Kosaka N, Konishi Y, et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat Commun 2014;5:3591. [Crossref] [PubMed]
- Chen Y, Xie Y, Xu L, et al. Protein content and functional characteristics of serum-purified exosomes from patients with colorectal cancer revealed by quantitative proteomics. Int J Cancer 2017;140:900-13. [Crossref] [PubMed]
- Li J, Sherman-Baust CA, Tsai-Turton M, et al. Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer. BMC Cancer 2009;9:244. [Crossref] [PubMed]
- Gabriel K, Ingram A, Austin R, et al. Regulation of the tumor suppressor PTEN through exosomes: a diagnostic potential for prostate cancer. PLoS One 2013;8:e70047 [Crossref] [PubMed]
- Khan S, Jutzy JM, Valenzuela MM, et al. Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS One 2012;7:e46737 [Crossref] [PubMed]
- Worst TS, von Hardenberg J, Gross JC, et al. Database-augmented Mass Spectrometry Analysis of Exosomes Identifies Claudin 3 as a Putative Prostate Cancer Biomarker. Mol Cell Proteomics 2017;16:998-1008. [Crossref] [PubMed]
- Kawakami K, Fujita Y, Matsuda Y, et al. Gamma-glutamyltransferase activity in exosomes as a potential marker for prostate cancer. BMC Cancer 2017;17:316-27. [Crossref] [PubMed]
- Kharaziha P, Chioureas D, Rutishauser D, et al. Molecular profiling of prostate cancer derived exosomes may reveal a predictive signature for response to docetaxel. Oncotarget 2015;6:21740-54. [Crossref] [PubMed]
- Logozzi M, Angelini DF, Iessi E, et al. Increased PSA expression on prostate cancer exosomes in in vitro condition and in cancer patients. Cancer Lett 2017;403:318-29. [Crossref] [PubMed]
- Muller L, Muller-Haegele S, Mitsuhashi M, et al. Exosomes isolated from plasma of glioma patients enrolled in a vaccination trial reflect antitumor immune activity and might predict survival. Oncoimmunology 2015;4:e1008347 [Crossref] [PubMed]
- Ueda K, Ishikawa N, Tatsuguchi A, et al. Antibody-coupled monolithic silica microtips for highthroughput molecular profiling of circulating exosomes. Sci Rep 2014;4:6232-40. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Gallo A, Tandon M, Alevizos I, et al. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One 2012;7:e30679 [Crossref] [PubMed]
- Eichelser C, Stuckrath I, Muller V, et al. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 2014;5:9650-63. [Crossref] [PubMed]
- Dejima H, Iinuma H, Kanaoka R, et al. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncology Letters 2017;13:1256-63. [Crossref] [PubMed]
- Liu Q, Yu Z, Yuan S, et al. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget 2017;8:13048-58. [PubMed]
- Wu H, Zhou J, Mei S, et al. Circulating exosomal microRNA-96 promotes cell proliferation, migration and drug resistance by targeting LMO7. J Cell Mol Med 2017;21:1228-36. [Crossref] [PubMed]
- Giallombardo M, Chacártegui Borrás J, Castiglia M, et al. Exosomal miRNA Analysis in Non-small Cell Lung Cancer (NSCLC) Patients' Plasma Through qPCR: A Feasible Liquid Biopsy Tool. J Vis Exp 2016;(111).
- Rabinowits G, Gercel-Taylor C, Day JM, et al. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 2009;10:42-6. [Crossref] [PubMed]
- Samsonov R, Burdakov V, Shtam T, et al. Plasma exosomal miR-21 and miR-181a differentiates follicular from papillary thyroid cancer. Tumour Biol 2016;37:12011-21. [Crossref] [PubMed]
- Chiam K, Wang T, Watson DI, et al. Circulating Serum Exosomal miRNAs As Potential Biomarkers for Esophageal Adenocarcinoma. J Gastrointest Surg 2015;19:1208-15. [Crossref] [PubMed]
- Warnecke-Eberz U, Chon SH, Holscher AH, et al. Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: comparison of miRNA profiles of exosomes and matching tumor. Tumour Biol 2015;36:4643-53. [Crossref] [PubMed]
- Tanaka Y, Kamohara H, Kinoshita K, et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer 2013;119:1159-67. [Crossref] [PubMed]
- Monzo M, Santasusagna S, Moreno I, et al. Exosomal microRNAs isolated from plasma of mesenteric veins linked to liver metastases in resected patients with colon cancer. Oncotarget 2017;8:30859-69. [Crossref] [PubMed]
- Tsukamoto M, Iinuma H, Yagi T, et al. Circulating Exosomal MicroRNA-21 as a Biomarker in Each Tumor Stage of Colorectal Cancer. Oncology 2017;92:360-70. [Crossref] [PubMed]
- Liu C, Eng C, Shen J, et al. Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget 2016;7:76250-60. [PubMed]
- Uratani R, Toiyama Y, Kitajima T, et al. Diagnostic Potential of Cell-Free and Exosomal MicroRNAs in the Identification of Patients with High-Risk Colorectal Adenomas. PLoS One 2016;11:e0160722 [Crossref] [PubMed]
- Matsumura T, Sugimachi K, Iinuma H, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer 2015;113:275-81. [Crossref] [PubMed]
- Ogata-Kawata H, Izumiya M, Kurioka D, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One 2014;9:e92921 [Crossref] [PubMed]
- Yan S, Han B, Gao S, et al. Exosome-encapsulated microRNAs as circulating biomarkers for colorectal cancer. Oncotarget 2017;8:60149-58. [PubMed]
- Jiao C, Jiao X, Zhu A, et al. Exosomal miR-34s panel as potential novel diagnostic and prognostic biomarker in patients with hepatoblastoma. J Pediatr Surg 2017;52:618-24. [Crossref] [PubMed]
- Sohn W, Kim J, Kang SH, et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med 2015;47:e184 [Crossref] [PubMed]
- Wang H, Hou L, Li A, et al. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res Int 2014;2014:864894 [PubMed]
- Lai X, Wang M, McElyea SD, et al. A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett 2017;393:86-93. [Crossref] [PubMed]
- Que R, Ding G, Chen J, et al. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J Surg Oncol 2013;11:219-27. [Crossref] [PubMed]
- Li Z, Ma YY, Wang J, et al. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther 2015;9:139-48. [PubMed]
- Li M, Rai AJ, DeCastro GJ, et al. An optimized procedure for exosome isolation and analysis using serum samples: Application to cancer biomarker discovery. Methods 2015;87:26-30. [Crossref] [PubMed]
- Huang X, Yuan T, Liang M, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol 2015;67:33-41. [Crossref] [PubMed]
- Manier S, Liu CJ, Avet-Loiseau H, et al. Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood 2017;129:2429-36. [Crossref] [PubMed]
- Asano M, Umezu T, Katagiri S, et al. Up-regulated exosomal miRNA-140-3p in CML patients with musculoskeletal pain associated with discontinuation of tyrosine kinase inhibitors. Int J Hematol 2017;105:419-22. [Crossref] [PubMed]
- Hornick NI, Huan J, Doron B, et al. Serum Exosome MicroRNA as a Minimally-Invasive Early Biomarker of AML. Sci Rep 2015;5:11295. [Crossref] [PubMed]
- Zhang L, Pan L, Xiang B, et al. Potential role of exosome-associated microRNA panels and in vivo environment to predict drug resistance for patients with multiple myeloma. Oncotarget 2016;7:30876-91. [PubMed]
- Pfeffer SR, Grossmann KF, Cassidy PB, et al. Detection of Exosomal miRNAs in the Plasma of Melanoma Patients. J Clin Med 2015;4:2012-27. [Crossref] [PubMed]
- Wang J, Zhou Y, Lu J, et al. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med Oncol 2014;31:148. [Crossref] [PubMed]
- Xu JF, Wang YP, Zhang SJ, et al. Exosomes containing differential expression of microRNA and mRNA in osteosarcoma that can predict response to chemotherapy. Oncotarget 2017;8:75968-78. [PubMed]
- Sugimachi K, Matsumura T, Hirata H, et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer 2015;112:532-8. [Crossref] [PubMed]
- Zhang J, Li S, Li L, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 2015;13:17-24. [Crossref] [PubMed]
- Schwarzenbach H, da Silva AM, Calin G, et al. Data Normalization Strategies for MicroRNA Quantification. Clin Chem 2015;61:1333-42. [Crossref] [PubMed]
- Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet 2014;15:423-37. [Crossref] [PubMed]
- Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014;25:666-81. [Crossref] [PubMed]
- Ahadi A, Brennan S, Kennedy PJ, et al. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci Rep 2016;6:24922. [Crossref] [PubMed]
- Qu L, Ding J, Chen C, et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell 2016;29:653-68. [Crossref] [PubMed]
- Pan L, Liang W, Fu M, et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol 2017;143:991-1004. [Crossref] [PubMed]
- Li Q, Shao Y, Zhang X, et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol 2015;36:2007-12. [Crossref] [PubMed]
- Dong L, Lin W, Qi P, et al. Circulating Long RNAs in Serum Extracellular Vesicles: Their Characterization and Potential Application as Biomarkers for Diagnosis of Colorectal Cancer. Cancer Epidemiol Biomarkers Prev 2016;25:1158-66. [Crossref] [PubMed]
- Liu T, Zhang X, Gao S, et al. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget 2016;7:85551-63. [Crossref] [PubMed]
- Zhang R, Xia Y, Wang Z, et al. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun 2017;490:406-14. [Crossref] [PubMed]
- Isin M, Uysaler E, Ozgur E, et al. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front Genet 2015;6:168. [PubMed]
- Li T, Xie J, Shen C, et al. Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res 2015;75:3181-91. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777 [Crossref] [PubMed]
- Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res 2016;6:1167-76. [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Weng W, Wei Q, Toden S, et al. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res 2017;23:3918-28. [Crossref] [PubMed]
- Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res 2013;73:5609-12. [Crossref] [PubMed]
- Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015;25:981-4. [Crossref] [PubMed]
- Dou Y, Cha DJ, Franklin JL, et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep 2016;6:37982. [Crossref] [PubMed]
- Hong BS, Cho JH, Kim H, et al. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics 2009;10:556. [Crossref] [PubMed]
- Hao YX, Li YM, Ye M, et al. KRAS and BRAF mutations in serum exosomes from patients with colorectal cancer in a Chinese population. Oncol Lett 2017;13:3608-16. [Crossref] [PubMed]
- Provencio M, Rodriguez M, Cantos B, et al. mRNA in exosomas as a liquid biopsy in non-Hodgkin Lymphoma: a multicentric study by the Spanish lymphoma oncology group. Oncotarget 2017;8:50949-57. [Crossref] [PubMed]
- Kalluri R, LeBleu VS. Discovery of Double-Stranded Genomic DNA in Circulating Exosomes. Cold Spring Harb Symp Quant Biol 2016;81:275-80. [Crossref] [PubMed]
- Cai J, Han Y, Ren H, et al. Extracellular vesicle-mediated transfer of donor genomic DNA to recipient cells is a novel mechanism for genetic influence between cells. J Mol Cell Biol 2013;5:227-38. [Crossref] [PubMed]
- Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 2014;24:766-9. [Crossref] [PubMed]
- Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem 2014;289:3869-75. [Crossref] [PubMed]
- Yang S, Che SP, Kurywchak P, et al. Detection of mutant KRAS and TP53 DNA in circulating exosomes from healthy individuals and patients with pancreatic cancer. Cancer Biol Ther 2017;18:158-65. [Crossref] [PubMed]
- Allenson K, Castillo J, San Lucas FA, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol 2017;28:741-7. [PubMed]
- Fricke F, Lee J, Michalak M, et al. TGFBR2-dependent alterations of exosomal cargo and functions in DNA mismatch repair-deficient HCT116 colorectal cancer cells. Cell Commun Signal 2017;15:14. [Crossref] [PubMed]
- Tan A, De La Pena H, Seifalian AM. The application of exosomes as a nanoscale cancer vaccine. Int J Nanomedicine 2010;5:889-900. [PubMed]
- Pitt JM, Charrier M, Viaud S, et al. Dendritic cell-derived exosomes as immunotherapies in the fight against cancer. J Immunol 2014;193:1006-11. [Crossref] [PubMed]
- Marleau AM, Chen CS, Joyce JA, et al. Exosome removal as a therapeutic adjuvant in cancer. J Transl Med 2012;10:134. [Crossref] [PubMed]


