
Identifying drivers of metastasis; towards a systematic approach
Metastasis is incontrovertibly linked to poor cancer patient survival. Cancer treatments that inhibit metastasis are predicted to improve cancer patient outcomes by preventing cancer dissemination. However, to control metastasis in patients necessitates an understanding of the biological drivers of metastasis. Drivers of metastasis can be thought of as factors that promote tumour dissemination, required at least for a part of the metastatic process. Yet, presently specific molecular drivers of metastasis largely remain unidentified, and very little evidence for ubiquitous metastasis-specific driver gene mutations has been identified to date (1,2).
Metastasis is a complex process, and it is becoming clear that identifying metastasis drivers requires novel approaches (3). Systems biology incorporates modelling of complex biological systems at the molecular, cellular, tissue and organismal levels, and so might be a suitable approach to study metastasis (4).
Metastasis refers to the acquisition of new aggressive behaviours, or hallmark properties (5) of cancer cells during tumour growth, following the initial development of a primary tumour. Of the different cancer types, melanoma is particularly aggressive and has a high propensity to metastasize early in its growth, at a stage when relatively few cell divisions have occurred (6). Despite remarkable advances in new treatments for metastatic melanoma in the past few years, outcomes for melanoma patients remain relatively modest due to frequently little or no treatment response, or alternatively some degree of initial response followed by resistance to therapy and relapse, or commonly occurring adverse side-effects (7). Targeting specific metastasis drivers may provide a revolutionary approach to treating cancer, and could lead to major advances in improving patient outcomes.
In the article by Agrawal et al. (8) the authors have used a systems biology approach to investigate glycosylation changes in clinical melanoma tissue samples from patients. The overall hypothesis was that altered glycosylation patterns would be a potential driver of metastasis and would contribute to aggressive behaviour in melanoma. To address this hypothesis, the authors compared differences in glycoprotein glycosylation between matched primary and metastatic tumour tissues, and found an increase in core fucosylation (addition of a fucose residue in α1,6 linkage to the first GlcNAc of the oligosaccharide core) mediated by fucosyl transferase 8 (FUT8) in metastatic melanoma. Improved sensitivity in the screening technology using lectin microarrays for low amounts of fixed tissue starting material to detect altered glycosylation in a cohort of patient samples was a key technical advance, providing a possible starting point for this research in using clinical samples. The authors used matched primary and metastasis melanoma tissues from a total of 17 patients, i.e., 34 samples in total to carry out a lectin microarray screen. Primary melanomas are small in size (often less than 0.5 mm3), and most of the primary melanoma tissue specimen had already been used for diagnosis, leaving little tissue available for screening. Additionally, primary melanoma tissue is frequently formalin-fixed and paraffin-embedded (FFPE), meaning that although archival tissue may be available, it needs to be deparaffinized first, and extensive cross-linking could limit detection sensitivity.
The authors found that metastatic tumours exhibited higher levels of poly/multi-antennary-N-acetyl-lactosamine, α-2,6 sialic acids, and core fucose, and lower levels of α-1,2 fucose structures than the corresponding primary melanoma samples. Having identified these differences in the glycomic profiles in the patient tissue samples, the authors next correlated their lectin microarray data with publicly available transcriptomic data from TCGA, focusing on glycosyltransferase genes that would be predicted to be involved in the synthesis of glycan epitopes detected by the lectin and antibody probes present in the lectin microarray. Among the observed glycan changes, the metastatic melanomas exhibited higher expression levels of the core fucosyltransferase, FUT8, associated with core fucose, than did primary melanomas. Following this, the authors then went on to assess the effects of silencing particular glycogenes (glycosyltransferases) on cell invasion, the latter being a key property of tumour cells required for metastasis. Gene-silencing screens, using small interfering RNA (siRNA) mini-screens, were carried out on two melanoma cell lines (SkMel147 and WM3211), in which the selection of specific glycosyltransferase candidates to be knocked down had previously been identified as being either “pro-metastatic” or “anti-metastatic”. For example, changes in fucosylation affect key pro-metastatic processes such as adhesion and immune recognition. As a functional assay of invasiveness, trans-well invasion assays were incorporated into the screening methodology for the two cell lines, and once again FUT8 was identified as one of the candidate pro-invasion/pro-metastatic genes to be selected for subsequent studies. FUT8 is a fucosyltransferase that promotes core fucosylation (which is one of the most common types of glycosylation involving the attachment of a fucose residue to N-glycans, O-glycans and glycolipids) of target glycoproteins.
Following the identification of FUT8 as a candidate glycogene for inducing invasive behaviour in melanoma cells, the authors then demonstrated that FUT8 silencing decreased the extent of in vivo melanoma metastasis occurring in xenograft tumours, using two metastatic melanoma cell lines, 113/6-4L (4L) and SkMel147, both of which express endogenous FUT8 at high levels. In addition, the authors showed that FUT8 expression is required in established metastases of 4L cells for further metastatic dissemination, using in vivo assays in mice.
The authors went on to show that up-regulation of FUT8 expression in metastatic melanoma was mainly due to transcriptional activation, and that the FUT8 promoter was predominantly responsive to changes in transforming growth factor β (TGF-β)-induced factor homeobox 2 (TGIF2) protein levels. TGIF2 binding to the FUT8 promoter was enriched in accordance with high levels of promoter activity and FUT8 expression.
One hundred and fourteen glycoproteins were identified as being commonly expressed in all three of the 4L, SkMel147 and MeWo cell lines, and in which core-fucosylation was deregulated together with high FUT8 expression. Gene ontology enrichment analysis for cell migration and locomotion control mechanisms was used to identify neural cell adhesion molecule L1 (L1CAM) as being common to all three cell lines, along with another 12 glycosylated proteins, including integrins α4, 6, and V, integrin B1, ADAM10, laminins β1 (LAMB1) and γ1 (LAMC1), pro-low-density lipoprotein receptor-related protein 1 (LRP1), cell surface glycoprotein MUC18 (MCAM), plexin-A1, plexin B2, and neuropilin 2 (NRP2). L1CAM was identified as a key core fucosylated glycoprotein in melanoma that mediated the pro-invasive effects of FUT8. High levels of glycosylation in L1CAM protein had already been known to regulate cell attachment, invasion and migration in cancer, including melanoma progression (9). Core fucosylation in L1CAM was found to be required for promoting plasmin cleavage of L1CAM.
Thus, to summarize, using a systems-based approach to study glycomic changes and corresponding enzymes associated with melanoma metastasis in patient samples, the authors identified molecular changes in metastatic tumours, including altered glycosylation and elevated FUT8 expression. These changes were associated with upregulation of core fucosylation by FUT8, and importantly, extensive functional analysis of invasion and metastasis, both in vitro and in vivo using mouse models underpinned the authors’ assertion that FUT8-mediated core fucosylation acts as a driver of melanoma metastasis, while its inhibition suppressed invasion and tumour dissemination. Increased FUT8 expression in metastatic melanoma was transcriptionally mediated by TGIF2 binding. Adhesion molecule, L1CAM, was identified as a glycoprotein target of FUT8, and was enriched in cell migration proteins in metastatic melanomas, and core fucosylation was found to impact L1CAM cleavage and the ability of L1CAM to support melanoma invasion. Therefore, the authors overall suggest that FUT8 may represent a new potential therapeutic target for melanoma metastasis.
Aside from glycosylation enzymes identified by Agrawal et al. (8), abnormalities have been identified in the expression of dozens of genes associated with melanoma aggressiveness, and a number of these genes have been suggested as potential metastasis drivers (see Table 1). Transcriptional differences between primary and metastatic melanomas could subsequently lead to downstream molecular changes, some of which could be therapeutically targeted to inhibit metastasis in advanced cancer patients. However, does systems biology provide a suitable approach to confidently identify novel metastasis drivers, and if so, how can this process be efficiently exploited? But firstly, what exactly is a “novel metastasis driver”.
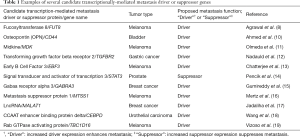
Full table
Carrying out systematic screening, using multiple “omic” platforms on matched primary and metastatic tumour samples, as well as combined functional analysis incorporating in vitro and in vivo studies, together with analysis of additional patient cohorts, and/or additional molecular characterization, is very likely to be the basis of future systems biology approaches, with the aim of identifying metastasis drivers. Nevertheless, despite present understanding of the term “driver mutation” in cancer, defining what is meant by a driver of metastasis is somewhat looser, especially since gene mutations, it would appear, constitute only a small part of the spectrum of possible driver molecular events in metastasis. To avoid potential confusion or unclear thinking in this field, it may be necessary to better define exactly what is meant by the term “driver” in the context of metastasis.
Recently, a potentially systematic approach outlined several criteria to define driver changes, which could be acquired in metastasis, and which would lead to changes in gene transcription in metastatic cancer cells (13,20). The driver changes debated involved the acquisition of driver epigenetic changes, but the same criteria would be applicable to transcriptional changes; accordingly several suggestions including considerations, approaches and models for the study of such metastasis drivers were made (13,20). Molecular changes may be identified in, and shared between many different metastatic tissue samples, and may be demonstrated to result in metastasis-related functional changes in tumour cells. It was asserted that these changes are more likely to be important in the context of metastasis than non-shared and/or functionally unrelated changes (13,20), which is broadly similar in concept to the frequency-based approaches used to identify cancer driver mutations (1).
Alterations showing a consistent enhancement, or reduction in their effect between matched metastatic versus primary tumour paired tissue samples, together with functional links to aggressive behaviour, and which are observed in multiple different metastatic samples, would fulfil several criteria that might be expected of candidate drivers of metastasis. Studies such as the one by Agrawal et al. (8) pave the way towards the establishment of a consensus on what constitutes metastasis drivers able to enhance the metastatic fitness of tumor cells (21), and therefore potential new therapeutic targets.
Acknowledgments
Funding: This work was supported by funding from the New Zealand Institute for Cancer Research Trust, and the Maurice Wilkins Centre for Molecular Biodiscovery.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Chen Qian (Center for Inflammation & Epigenetics, Houston Methodist Hospital Research Institute, Houston, TX, USA).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.09.52). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Gibson WJ, Hoivik EA, Halle MK, et al. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat Genet 2016;48:848-55. [Crossref] [PubMed]
- Lee JH, Zhao XM, Yoon I, et al. Integrative analysis of mutational and transcriptional profiles reveals driver mutations of metastatic breast cancers. Cell Discov 2016;2:16025. [Crossref] [PubMed]
- Likic VA, McConville MJ, Lithgow T, et al. Systems biology: the next frontier for bioinformatics. Adv Bioinformatics 2010:268925.
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Shain AH, Bastian BC. From melanocytes to melanomas. Nat Rev Cancer 2016;16:345-58. [Crossref] [PubMed]
- Luke JJ, Flaherty KT, Ribas A, et al. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat Rev Clin Oncol 2017;14:463-82. [Crossref] [PubMed]
- Agrawal P, Fontanals-Cirera B, Sokolova E, et al. A Systems Biology Approach Identifies FUT8 as a Driver of Melanoma Metastasis. Cancer Cell 2017;31:804-19.e7. [Crossref] [PubMed]
- Altevogt P, Doberstein K, Fogel M. L1CAM in human cancer. Int J Cancer 2016;138:1565-76. [Crossref] [PubMed]
- Ahmed M, Sottnik JL, Dancik GM, et al. An Osteopontin/CD44 Axis in RhoGDI2-Mediated Metastasis Suppression. Cancer Cell 2016;30:432-43. [Crossref] [PubMed]
- Olmeda D, Cerezo-Wallis D, Riveiro-Falkenbach E, et al. Whole-body imaging of lymphovascular niches identifies pre-metastatic roles of midkine. Nature 2017;546:676-80. [Crossref] [PubMed]
- Nadauld LD, Garcia S, Natsoulis G, et al. Metastatic tumor evolution and organoid modeling implicate TGFBR2 as a cancer driver in diffuse gastric cancer. Genome Biol 2014;15:428. [Crossref] [PubMed]
- Chatterjee A, Stockwell PA, Ahn A, et al. Genome-wide methylation sequencing of paired primary and metastatic cell lines identifies common DNA methylation changes and a role for EBF3 as a candidate epigenetic driver of melanoma metastasis. Oncotarget 2017;8:6085-101. [PubMed]
- Pencik J, Schlederer M, Gruber W, et al. STAT3 regulated ARF expression suppresses prostate cancer metastasis. Nat Commun 2015;6:7736. [Crossref] [PubMed]
- Gumireddy K, Li A, Kossenkov AV, et al. The mRNA-edited form of GABRA3 suppresses GABRA3-mediated Akt activation and breast cancer metastasis. Nat Commun 2016;7:10715. [Crossref] [PubMed]
- Mertz KD, Pathria G, Wagner C, et al. MTSS1 is a metastasis driver in a subset of human melanomas. Nat Commun 2014;5:3465. [Crossref] [PubMed]
- Jadaliha M, Zong X, Malakar P, et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget 2016;7:40418-36. [Crossref] [PubMed]
- Wang YH, Wu WJ, Wang WJ, et al. CEBPD amplification and overexpression in urothelial carcinoma: a driver of tumor metastasis indicating adverse prognosis. Oncotarget 2015;6:31069-84. [Crossref] [PubMed]
- Vizoso M, Ferreira HJ, Lopez-Serra P, et al. Epigenetic activation of a cryptic TBC1D16 transcript enhances melanoma progression by targeting EGFR. Nat Med 2015;21:741-50. [Crossref] [PubMed]
- Chatterjee A, Rodger EJ, Eccles MR. Epigenetic drivers of tumourigenesis and cancer metastasis. Semin Cancer Biol 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Patel SA, Vanharanta S. Epigenetic determinants of metastasis. Mol Oncol 2016; [Epub ahead of print]. [PubMed]


