
Mighty RapaLink-1 vanquishes undruggable mutant mTOR in glioblastoma
Recent emergence of a third generation of mechanistic target of rapamycin (mTOR) inhibitors, termed RapaLink-1, have generated hope in treatment of glioblastoma, the most aggressive primary brain tumor in adults. Produced by coupling first and second generation mTOR inhibitors (1), RapaLink-1 displays superior efficacy in inhibition of the mTOR complex I (mTORC1) pathway. This led to the inhibition of proliferation with relatively longer durability in glioblastoma cells and xenograft models. Importantly, RapaLink-1 crosses the blood-brain barrier and effectively targets mutant mTOR glioblastoma cells. RapaLink-1 is a bivalent inhibitor generated by combining the analogue inhibitor of mTOR, rapamycin, with the ATP-competitive inhibitor of mTOR, MLN0128. The resulting compound is more potent than first- and second-generation mTOR inhibitors. Investigation by Fan et al. [2017] (1) examined the effect of RapaLink-1 and RapaLink-2 on glioblastoma cell growth and proliferation by targeting the mTORC1.
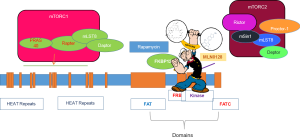
mTOR is a member of the phosphoinositide 3-kinase-related kinase (PI3K) family with homologs in all eukaryotes (2,3). The N-terminus of mTOR contains several huntingtin elongation factor 3 protein phosphatase 2A TOR1 (HEAT) repeats to facilitate the majority of interactions with other proteins. The FKB-FRAP domain that directly binds to rapamycin is located at the C-terminus of the mTOR protein. The C-terminus also contains a kinase domain that places it in the PI3K family. The canonical PI3K/Akt/mTOR signaling axis regulates various physiological functions including cellular growth, proliferation, differentiation, and metabolism (2,4,5). Hyperactivation of mTOR signaling in cancers occurs via multiple mechanisms (6) and results in an increase in cell growth, causing some cell types to enter the cell cycle. Aberrant mTOR pathway activity is prevalent in glioblastoma, leading to abnormalities in cell proliferation and motility, thereby resulting in uncontrolled growth and dissemination (7,8). Mutations of PTEN are found in approximately 70–90% of glioblastoma that leads to deregulation of the PI3K/Akt/mTOR signaling axis. In addition to frequent deletions and mutations of the tumor suppressor PTEN, about 86% of the glioblastoma samples display at least one genetic event leading to activation of the RTK/PI3K pathway (9). Constitutive activation of mTOR via single point mutations has been shown in many cancers, including glioblastoma (10). Hyper-activation of mTOR signaling makes it an attractive target for therapeutic intervention. In fact, first-generation mTOR inhibitors, rapamycin, and its analogue, rapalogs, exclusively bind to the component of mTORC1 and inhibit mTOR activity with limited success in clinical settings. Among these agents, first-generation mTOR inhibitors, rapamycin, and its analogue, rapalogs, inhibit mTOR by forming a complex with the immunophilin FKBP12. FKBP12 then exclusively binds directly to the component of mTORC1 (4). Second generation ATP-competitive inhibitor of mTOR shows potent and selective inhibition of mTORC1 and mTORC2. It also shows anti-proliferative activity, however, it is limited in suppressing mTORC1. Resistant glioblastoma cells as well as therapy related formation of mutant mTOR were the major obstacles of mTOR inhibitors.
Creation of RapaLink, the third generation of mTOR complex inhibitors, was achieved by linking first generation analogue binding inhibitor, rapamycin, to the second generation inhibitor, MLN0128, hence termed, RapaLink (Figure 1). Two forms of RapaLink were created: RapaLink-1 and Rapalink-2, which differ in size of linker length. RapaLink-1was superior in inhibiting mTOR complex as it potently reduced levels of both p-4EBP1, an important component of mTORC1, and cell proliferation as compared with RapaLink-2. RapaLink-1 was shown to be more potent in inhibiting glioblastoma cell growth inhibition as well as halting cells in G0/G1 of the cell cycle as compared to rapamycin or MLN0128 alone. As expected, rapamycin suppressed the mTORC1 target, p-RPS6S235/236, and MLN0128 inhibited both the mTORC1 targets p-RPS6S235/236 and p-4EBP1T37/46, as well as mTORC2 targets p-AKTS473 in a dose-dependent manner. In this investigation, Fan et al. [2017] (1) demonstrated that RapaLink-1 selectively inhibited p-RPS6S235/236 and p-4EBP1T37/46 at a low dose of 1.56 nM. Suppression of mTORC2 was achieved at higher doses only in two human glioblastoma cell lines, LN229 and U87MG, and in short-term primary cultures from patient-derived xenografts. It is important to note that glioblastoma may develop new driver mutations in response to therapies, including activating mutations in mTOR itself. Crucially, RapaLink-1 effectively reduced cell proliferation of glioblastoma cells expressing either wild-type or activating mutation of mTOR, namely mTORR2505P and mTORS2115Y. Rapamycin treatment also caused a significant suppression in cell growth irrespective of mTOR status, albeit to a lesser extent as compared to RapaLink-1. On the other hand, MLN0128 treatment was less effective in cells expressing mutant mTOR. Fan et al. [2017] (1) further demonstrated that RapaLink-1 blocked activation of p-4EBP1T37/46 regardless of mutational status of mTOR. In contrast, levels of p-4EBP1T37/46 remained higher in mTOR mutant glioblastoma cells treated with rapamycin or MLN08. These results strengthened the effectiveness of RapaLink-1 as compared to first- and second-generation mTOR inhibitors.
Rapamycin and its chemically related compounds have recently been used in clinical trials for the treatment of cancer due to their inhibitory effects on the mTOR pathway (12). Clinical trials using rapamycin and its analogs (CCI-779/temsirolimus, RAD001/everolimus, and AP23573) have shown promising yet challenging results in the management of various tumors including glioblastoma. In fact, a trial by Cloughesy et al. [2008] (13) showed that rapamycin successfully reduced tumor growth in half of the patients with PTEN-deficient, recurrent glioblastoma. However, there were challenges in targeting the mTOR pathway as a significant number of patients showed an increased level of phosphorylated AKT and PRAS40. It was concluded that this activation could be due to loss of negative feedback, resulting in decreased time for progression free-survival in these patients, causing poor clinical outcomes (13). This particular trial stands out as it demonstrates the molecular mechanisms of glioblastoma developing resistance to rapamycin and its analogs. Specifically, this trial establishes the limited therapeutic effectiveness of mTOR inhibition with rapamycin and its analogs due to loss of negative feedback. Therefore, despite significant advancement in targeted therapy, glioblastoma remains incurable possibly in part due to activation of mitogenic pathways such as RAS/ERK1/2 (7,8). Preclinical and clinical studies of ATP-competitive mTOR inhibitors that target both mTORC1 and mTORC2 demonstrated greater effectiveness than rapalogs in treatment of cancer. However, it is important to consider that the acute mTORC1 inhibition can activate the negative feedback loop leading to triggering of PI3K/PDK1/Akt (Thr308) and IRS1 via mitogenic pathways, leading to relentless cell survival and proliferation (7,8).
In vivo studies performed in mTOR xenograft models using BALB/Cnu/nu mice, administered daily I.P. injections of vehicle, MLN0128, rapamycin, or RapaLink-1 every 5th day, showed that RapaLink-1 inhibited components of mTORC1, p-RPS6S235/236 and p-4EBP1T37/46 in a dose-dependent manner in the brain. Conversely, it failed to inhibit the mTORC2 substrate p-AKTS473in vivo. An important observation of this study was that RapaLink crosses through the blood-brain barrier (1). Furthermore, experiments revealing tumor burden, mTOR signaling, and survival analysis revealed that RapaLink-1 caused regression and subsequent stabilization of tumor size. In contrast vehicle, rapamycin, or MLN0128-treated mice displayed a steady growth in their tumor. Furthermore, RapaLink-1 blocked the expression of p-4EBP1T37/46 while MLN0128 or rapamycin had modest effects in reducing the levels of p-4EBP1T37/46. Suppression of p-RPS6S235/236 was achieved in all treatment groups individually, however inhibition of mTORC2 substrate p-AKTS473 was achieved only after treatment with MLN0128.
Further in this study, xenograft/mice-treated with inhibitors were followed for survival/analyses, which showed that RapaLink-1 treated animals had much improved survival as compared to vehicle, MLN012, or rapamycin-treated animals. In addition, proliferative index was robustly suppressed by RapaLink-1 treatment whereas it was only modestly suppressed by rapamycin or MLN0128 treatments. Authors further demonstrated that patient-derived glioblastoma cells in xenograft models displayed a marked reduction in tumor growth following IP administration of RapaLink-1 when analyzed by luciferase signal as compared with vehicle, rapamycin, or MLN0128 treatments (1). As with earlier treatments, RapaLink-1 efficiently blocked the expression of p-4EBP1T37/46 in tumors, while both MLN0128 and rapamycin only modestly reduced p-4EBP1T37/46. Levels of p-RPS6S235/236 remained indistinctly suppressed by all treatments and MLN0128 exclusively inhibited p-AKTS473. Structurally, binding of rapamycin to FKBP12 and FRAP is subjected to two different hydrophobic binding pockets which helps occupancy of rapamycin in cells leading to a sustained suppression of mTORC1 substrate, p-RPS6S235/236 (14). Fan et al. [2017] (1) showed that RapaLink-1 compared with both rapamycin and MLN0128 was most effective in suppressing human glioma cell growth for a long period of time and phosphorylation of RPS6 and 4EBP1 was down-regulated over a period of 24 h.
First and second generation inhibitors of mTOR displayed incomplete effects in glioblastoma tumors in both preclinical and clinical studies. Rapamycin and its analogues allosteric inhibitors inhibit mTORC1 by targeting S6K without inhibiting 4EBP1 (15). On the other hand, MLN0128 was more effective than rapamycin in in vitro studies, as it was able to inhibit mTORC1 target 4EBP1, but displayed shorter residence time (16). An improved efficacy of RapaLink-1 in in vivo studies may be attributed to its ability to efficiently block 4EBP1 as well as its prolonged residence time. Furthermore, in glioblastoma, 4EBP1is considered as a more reliable marker for mTORC1 activity and therefore inhibition of 4EBP1 is crucial in suppressing mTOR activity (17). RapaLink-1 treated cells showed higher levels of FKBP12 bound compared with cells treated with rapamycin. The rapamycin-FKBP12 complex binds only to FRB, whereas the RapaLink-1-FKBP12 complex binds to FRB and to the mTOR kinase domain, thereby increasing affinity as well as stability leading to higher efficacy. Despite its size, RapaLink-1 was able to cross the blood-brain barrier and induce regression of tumor burden in orthotopic/xenograft models for brain cancer. This class of agents thus holds promise for future therapy of patients with glioblastoma.
While RapaLink-1 promoted regression in glioblastoma models, this initial regression was followed by regrowth of the tumor. Such recurrence is consistent with data suggesting that mTOR inhibitors as monotherapies are not sufficient to achieve anti-tumor responses in most cancers (6). Studies to establish the basis for recurrence, such as induction of autophagy, negative feedback loops, rewiring, or acquisition of resistance by emergence of novel mutations will determine the means that promote apoptosis and overcome resistance to position RapaLink-1 for clinical development in treatment of glioblastoma.
The mTORC1 complex is composed of proteins such as regulatory associated protein of mTOR (RAPTOR), which is sensitive to rapamycin treatment. It has been shown that mTORC1 function is tightly regulated by PI3K/AKT. In contrast to mTORC1, the mTORC2 complex is sensitive to growth factors but not nutrients, and is associated with the rapamycin-insensitive companion of mTOR (RICTOR) along with other proteins (11). The major distinguishing characteristic of the mTORC1 and mTORC2 is their differential sensitivity to rapamycin (5). mTORC1 regulates protein synthesis through phosphorylating its downstream substrates, 4EBP1 (also called EIF4EBP1) and p70 S6K1/2 (11). The mTOR-dependent phosphorylation of p70 S6K1/2 promotes translation initiation as well as elongation and regulates cellular growth (11). mTORC2 modulates growth factor signaling by phosphorylating the C-terminal hydrophobic motif of some AGC kinases, such as AKT and SGK. Pharmacological inhibition of these complexes has been difficult to achieve. A phase I trial of rapamycin in PTEN-deficient glioblastoma patients showed some promising results with inherent difficulties of targeting mTOR pathways. A significant number of patients showed increased activated levels of pAKTSer473 following rapamycin treatment, which was correlated with shorter time to progression. The observed AKT activation was likely due to an alteration of signaling feedback loops, highlighting the complexity of prolonged rapamycin treatment.
Rapamycin (sirolimus) and its analogs, RAD001 (everolimus) and CCI-779 (temsirolimus), suppress mTOR activity through an allosteric mechanism that acts at a distance from the ATP binding catalytic site. Major disadvantages of rapamycin and other related compounds are that it suppresses TORC1 mediated S6K activation, thereby blocking a negative feedback loop. This led to the activation of PI3K/AKT and Ras/MEK/ERK signaling pathways, promoting cell survival and growth. Rapamycin remains an incomplete inhibitor of mTORC1 (18). In recent years, novel small ATP binding site molecules have been identified that directly inhibit mTOR, unlike rapamycin, which is an allosteric inhibitor of mTOR. In addition, novel ATP binding compounds with pyrazolopyrimidines are shown to inhibit members of the PI3K family, including mTOR. One such compound, PP242, is an ATP-competitive inhibitor of mTOR, which shows potent and selective inhibition of mTORC1 and mTORC2 (19). These molecules are often phrased “TORKinibs” for their ability to inhibit TOR kinase. First-generation TORi compounds such as PP242 inhibits mTORC1/C2 in AKT/mTOR signaling. Novel MLN0128, a second-generation of TORi compound (20), appears to be more potent.
Limitations of rapalogs-based clinical strategies have pushed toward the development of a second generation of mTOR inhibitors known as ATP-competitive mTOR kinase inhibitors (TORKIs), which target the kinase domain of mTOR and inhibit its catalytic activity. From a mechanistic point of view, the advantage of these drugs relies on the ability of inhibiting the kinase activity of both the TORC1 and TORC2 complexes while also blocking the feedback activation of PI3K/Akt signaling (18,21,22). Combinatory strategies may provide a way to overcome such resistance and therefore improve efficacy of mTOR targeting agents in the clinical context (23). In addition, mTOR has also been involved in regulation of stem cells (3).
The development of a third generation of mTOR inhibitors, such as RapaLink-1, holds the potential to overcome the resistance to the rapaloge or TORKi, perhaps due to its ability to bind to two drug-binding pockets of mTOR (Figure 1). As demonstrated in breast cancer cells, RapaLink-1 has the ability to target MCF-7 cells that carried three somatic mutations within mTOR, FRB-FKBP1, or Kinase domain conferring resistance (24). It is important to realize that the hyperactive M2327I mutation as seen here, similar to other mTOR kinase domain mutations, are seen in patients with no prior treatments (25), which may influence the sensitivity to ATP-competitive mTOR inhibitors. While FRB-domain mutations can cause resistance to therapy by disrupting the interaction of FKBP12-rapamycin complex to mTOR, influencing the binding of the inhibitor, the Kinase domain mutations cause hyperactivation of the kinase activity. Marked by such challenges, a drug-modeling technique has created a novel bivalent mTOR inhibitor such as Rapalink. Rapalink consists of a rapamycin-FRB-binding compound linked to a TORKi, which can target both FRB-domain as well as Kinase domain of mTOR. RapaLink potently inhibits the mTORC1 pathway by inhibiting the phosphorylation of 4EBP1 and growth inhibition both in vitro and in vivo, at levels comparable to rapamycin or a combination of rapamycin with MLN0128. Importantly, RapaLink-1also targets cells expressing specific mTOR mutations.
In summary, inhibition of the mTOR pathway following bivalent kinase inhibitor has proven to be therapeutic for glioblastoma and future clinical studies are expected to reveal the significance of such unique approaches. It is not unreasonable to envision that such therapeutic options may be effective in successfully treating patients with hyperactive mTOR kinase domain mutations and/or those who have developed resistance to rapalogs or ATP-competitive inhibitors. Future clinical trials may suggest whether RapaLink-1 results in therapy related resistance or activates alternate pathways due to the loss of negative feedback. Nevertheless, it is worth noting that bivalent kinase inhibitors such as Rapalink-1 are unique in design as described by Fan et al. (1) to provide more stable and potent suppressive effects on mTOR pathways in glioblastoma. Therefore, studies such as those presented here using novel bivalent inhibitors may serve as the initial foundation for developing innovative strategies in treatment of fatal cancers such as glioblastoma.
Acknowledgments
Funding: Supported by funds from Advanced Research Foundation.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Ning Huang (Department of Neurosurgery, The Second Affiliated Hospital of Chongqing Medical University, Chongqing, China).
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.09.36). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fan Q, Aksoy O, Wong RA, et al. A Kinase Inhibitor Targeted to mTORC1 Drives Regression in Glioblastoma. Cancer Cell 2017;31:424-35. [Crossref] [PubMed]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012;149:274-93. [Crossref] [PubMed]
- Russell RC, Fang C, Guan KL. An emerging role for TOR signaling in mammalian tissue and stem cell physiology. Development 2011;138:3343-56. [Crossref] [PubMed]
- Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci Signal 2009;2:pe24. [Crossref] [PubMed]
- Jacinto E, Loewith R, Schmidt A, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 2004;6:1122-8. [Crossref] [PubMed]
- Ilagan E, Manning BD. Emerging role of mTOR in the response to cancer therapeutics. Trends Cancer 2016;2:241-51. [Crossref] [PubMed]
- Jhanwar-Uniyal M, Jeevan D, Neil J, et al. Deconstructing mTOR complexes in regulation of Glioblastoma Multiforme and its stem cells. Adv Biol Regul 2013;53:202-10. [Crossref] [PubMed]
- Jhanwar-Uniyal M, Amin AG, Cooper JB, et al. Discrete signaling mechanisms of mTORC1 and mTORC2: Connected yet apart in cellular and molecular aspects. Adv Biol Regul 2017;64:39-48. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061-8. [Crossref] [PubMed]
- Sato A, Sunayama J, Matsuda K, et al. Regulation of neural stem/progenitor cell maintenance by PI3K and mTOR. Neurosci Lett 2010;470:115-20. [Crossref] [PubMed]
- Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer 2006;6:729-34. [Crossref] [PubMed]
- Sokolosky ML, Stadelman KM, Chappell WH, et al. Involvement of Akt-1 and mTOR in sensitivity of breast cancer to targeted therapy. Oncotarget 2011;2:538-50. [Crossref] [PubMed]
- Cloughesy TF, Yoshimoto K, Nghiemphu P, et al. Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med 2008;5:e8 [Crossref] [PubMed]
- Choi J, Chen J, Schreiber SL, et al. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 1996;273:239-42. [Crossref] [PubMed]
- Baretić D, Williams RL. The structural basis for mTOR function. Semin Cell Dev Biol 2014;36:91-101. [Crossref] [PubMed]
- Bradshaw TJ, Bowen SR, Deveau MA, et al. Molecular imaging biomarkers of resistance to radiation therapy for spontaneous nasal tumors in canines. Int J Radiat Oncol Biol Phys 2015;91:787-95. [Crossref] [PubMed]
- Fan QW, Nicolaides TP, Weiss WA. Inhibiting 4EBP1 in glioblastoma. Clin Cancer Res 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Thoreen CC, Sabatini DM. Rapamycin inhibits mTORC1, but not completely. Autophagy 2009;5:725-6. [Crossref] [PubMed]
- Feldman ME, Apsel B, Uotila A, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol 2009;7:e38 [Crossref] [PubMed]
- Zeng Z, Wang RY, Qiu YH, et al. MLN0128, a novel mTOR kinase inhibitor, disrupts survival signaling and triggers apoptosis in AML and AML stem/ progenitor cells. Oncotarget 2016;7:55083-97. [Crossref] [PubMed]
- Neil J, Shannon C, Mohan A, et al. ATP-site binding inhibitor effectively targets mTORC1 and mTORC2 complexes in glioblastoma. Int J Oncol 2016;48:1045-52. [PubMed]
- Thoreen CC, Kang SA, Chang JW, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 2009;284:8023-32. [Crossref] [PubMed]
- Feldman ME, Shokat KM. New inhibitors of the PI3K-Akt-mTOR pathway: insights into mTOR signaling from a new generation of Tor Kinase Domain Inhibitors (TORKinibs). Curr Top Microbiol Immunol 2010;347:241-62. [Crossref] [PubMed]
- Rodrik-Outmezguine VS, Okaniwa M, Yao Z, et al. Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature 2016;534:272-6. [PubMed]
- Grabiner BC, Nardi V, Birsoy K, et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov 2014;4:554-63. [Crossref] [PubMed]


