Potential of circulating nucleosome-associated histone modifications in cancer
Introduction
Cancer has traditionally been considered as a genetic disease. However, accumulating evidence revealed that epigenetic mechanisms contribute substantially to the high complexity of cancer development and progression (1). Epigenetics is defined as the study of gene expression that is not encoded by the DNA sequence and mainly refers to alterations in DNA methylation, histone modifications, and non-coding RNAs (2). These mechanisms regulate the high complexity of the mammalian genome and affect cell proliferation, differentiation, and cellular homeostasis. During the past decade, the research into the role of posttranslational modifications of histone proteins (PTHMs) in chromatin organization and gene expression has expanded (3). Enzymatic machinery that establishes PTHMs is often deregulated in cancer and altered patterns of PTHMs have been defined for various cancers (4). Nucleosomes constitute complexes of histones and DNA and may be released from cells into the blood circulation during cell death processes. They could be valuable sources for the detection of cancer-related alterations of PTHMs in bodily fluids that can be utilized in cancer detection, diagnosis, treatment evaluation or prognosis (5). As such biomarkers offer many advantages including minimally invasiveness and easy accessibility (6), scientific interest in circulating nucleosomes and PTHMs as relevant parts in the field of “liquid biopsy” is growing. In this review we provide a short insight into PTHMs and outline published data revealing their potential as circulating cancer biomarkers.
Posttranslational histone modifications
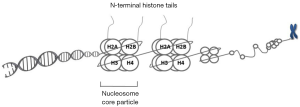
Eukaryotic chromatin has a compact organization composed of DNA, histones and the non-histone proteins. Non-histone proteins are less abundant than histones and more variable between tissues and species and include mainly scaffold proteins, DNA polymerase, heterochromatin protein 1 and polycomb proteins. Histones are primary protein components of chromatin and include small and highly conserved proteins H1, H2A, H2B, H3, and H4 (7). The basic structural repeating unit of chromatin is the nucleosome core particle that contains two copies of core histone proteins (H2A, H2B, H3 and H4) and 147-bp DNA wrapped around this histone octamer (Figure 1). H1 protein binds to the “linker DNA” region between nucleosomes.
The core histones possess highly dynamic N-terminal amino acid tails extending from the surface of nucleosome. These tails are subject to a variety of posttranslational modifications (8). Several different types of PTHMs have been identified in eukaryotic cells where in histone acetylation and methylation as ubiquitous marks of chromatin are key players of gene regulation. These have been mostly implicated in cancer development and progression (9). Acetylation of histone tails is typically associated with transcriptional activation of genes as this mark changes net positive charge of the histone proteins, providing access to DNA sequence information (10). The acetylation status of the histones is determined by a dynamic balance between histone acetyltransferases (HATs) and histone deacetylases (HDACs). By removing acetyl groups, HDACs reverse histone acetylation and affect the expression of many cancer critical genes. Aberrant expression of HDACs has been linked to a variety of malignancies (11). They are supposed to be involved in multiple stages of cancer and in most cases; a high level of HDACs is associated with advanced disease and poor outcomes in patients (11). Thus HDACs represent a relevant target of cancer therapeutics.
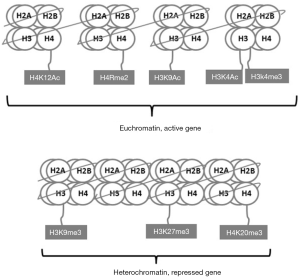
Unlike HATs and HDACs, histone methyltransferases (HMTs) and histone demethylases (HDMs) work in a specific manner to recognize and modify distinct basic target amino acid residues (12). Histones are methylated by addition of one to three methyl groups to the side chains of lysine or arginine residues by histone lysine methyltransferases (HKMTs) and protein arginine methyltransferases (PRMTs), respectively. HKMTs consist of two main classes, the SET domain containing family and the DOT1 family (13). The functional consequences of methylation depend mainly on the number of methyl groups and their location within the histone tail (14). For instance, histone 3 lysine 4 di- and trimethylation (H3K4me2 and H3K4me3, respectively) and histone 3 lysine 9 monomethylation (H3K9me1) are examples for modifications that are associated with open chromatin and active gene expression. In contrast, histone 3 lysine 27 di- and trimethylation (H3K27me2 and H3K27me3, respectively) are linked with gene repression. Histone 3 lysine 9 trimethylation (H3K9me3) and histone 4 lysine 20 trimethylation (H4K20me3) are two repressive epigenetic marks that are enriched in pericentric heterochromatin and important mediators of genomic stability (Figure 2) (15). Similar to HATs and HMTS, the dysregulation of HMTs and HDMs results in aberrant histone modification patterns in cancer cells (13). Several studies linked global changes of methylation marks to prognosis of patients with different types of cancer (16-18).
Techniques for studying histone modifications
Various techniques are available to study the function, abundance or interacting partners of PTHMs. Antibody-based approaches such as enzyme-immunoassays or Western Blots are broadly applied to detect PTHMs. One should be aware that the quality and specificity of antibodies being used is a major matter of the histone research (19). Cross-reactivity with similar PTHMs (such as double or triple methylation), with alternative histone modification sites or with other nuclear proteins represent the most obvious obstacles in these assays. For the detection of PTHMs by Western blotting, whole-cell lysates are often used (20). For some applications, it may be necessary to enrich or purify histone proteins. This approach provides, however, no quantitative data. Enzyme linked immunosorbent assays (ELISAs) are another antibody based technique at low cost and easy to be used that allows to quantify the number of histone marks in cell lysates or bodily fluids. Current research reveals a relatively high reproducibility of PTHMs detection by this approach (21,22). The number of studies that have measured PTHMs in serum or plasma using ELISA-based assays is accumulating (22-26). Chromatin immunoprecipitation (ChIP) is another approach to employ specific antibodies. The strength of this technique is the identification of genomic sites that are enriched for a particular histone modification. For this purpose, immunoprecipitated DNA is applied in subsequent real-time PCR to determine histone marks at given loci. Large-scale enrichment analysis can also be performed using a variety of massive parallel DNA sequencing (ChIP-seq) methods to identify the distribution of PTHMs genome-wide (27). Our research revealed the detection of circulating nucleosome-associated histone marks in blood circulation by using ChIP assays (28-30). It is, however, to note that sample preparation for ChIP and ChIP-seq assays includes multiple steps which are critical to the success of the experiments and affect the reproducibility, bias, and sensitivity of the technique.
Another elegant technique applied in histone research is mass spectrometry that proved to be effective in identifying and quantifying PTHMs and their binding proteins, beyond the limitations of antibody use (31,32). A major advantage of mass spectrometry-based methods is their capability to characterize combinatorial histone PTHMs simultaneously occurring on the same molecule (33). However, higher workload and sensitivity issues for the detection of specific PTHMs have to be considered. Nevertheless, mass spectrometry may be superior to immunoassays in order to provide the specific and comprehensive quantification of PTHM panels. In a first report employing mass spectrometry Fraga et al. (34) have described global loss of histone H4 trimethylation and acetylation as a common hallmark of human cancer cells. Since then numerous studies have applied this technique to characterize PTHMs in clinical samples (34,35).
Sample preparation for liquid biopsy
Pre-analytical variables may affect the specificity and sensitivity of detection of circulating genetic and epigenetic markers (36). A crucial step is blood processing and storage temperature that may influence marker stability and concentration (37). Ethylenediaminetetraacetic acid (EDTA) is the most used anticoagulant to stabilize blood during the time between sample drawn and processing. In order to get rid of contaminating cells, both filtration and repeated centrifugations of plasma or serum were found to be useful (38). It has been reported that circulating nucleosomes are relatively stable on long-term storage of sera at −70 °C (39). Similarly, pre-analytical variables such as contaminating cells, within-day variation, varying time before centrifugation had no significant influence on the level of histone methylation in circulating nucleosomes measured by an ELISA-based assay (25).
Clinical impact of circulating nucleosomes
Cells dying from necrosis or apoptosis are considered to be the main sources of extracellular nucleosomes (40,41). It is also assumed that active release from living cells contributes to some part of circulating nucleosomes (42). Circulating nucleosomes are shown predominantly as mono- or oligonucleosome (43). By deep sequencing cell-free DNA (cfDNA) from blood plasma, it was found out in a recent study that the cfDNA nucleosome occupancies correlate well with the nuclear architecture, gene structure, and expression observed in cells, suggesting that they could inform the cell type of origin (44). Nucleosomes are stable structures in circulation (39) and can be detected by ELISA-based measurement in serum and plasma. In the case of cancer increased amounts of nucleosomes enter blood circulation due to a higher cellular turnover (45) and impaired clearance by macrophages (41). In a number of studies circulating nucleosomes have been investigated as a diagnostic marker. A first study measuring circulating nucleosomes was conducted in breast cancer and described elevated levels of circulating nucleosomes in patients compared with those of healthy controls (46). In a subsequent larger study including 418 patients with malignant tumors, 109 patients with benign diseases and 63 healthy individuals, it was reported that sera of patients with malignant tumors contained considerably higher amounts of nucleosomes compared with those of healthy individuals (47). However, comparing these results to levels in patients with many benign diseases, the difference was not statistically significant, reducing their clinical utility for detecting cancer.
As circulating nucleosomes are released in response to chemotherapeutic agents, few studies have utilized the quantification of circulating nucleosomes levels to predict tumor responses. Detection of patients not responding to therapy before or at an early phase of treatment regime would enable to modify the treatment regime and ultimately save patients from the systemic side-effects of ineffective chemotherapy (48). In patients with advanced lung cancer, pre-therapeutic levels of circulating nucleosomes were significantly lower in patients who responded to chemotherapy (49,50). Furthermore, these patients experienced a smaller increase and greater decrease in circulating nucleosomes following the start of treatment. Similar results were obtained by Fahmueller et al. when they analyzed the nucleosome levels in sera of patients with metastasized colorectal cancer (CRC) undergoing selective internal radiation therapy (SIRT). They found that high increases 24 hours after application of this therapy, indicated poor therapy response and reduced survival time (51). Also Yörüker et al. (52) reported that the patients with CRC who had distant metastasis have higher nucleosome levels supporting the hypothesis that circulating nucleosomes provide predictive and prognostic information in diverse cancer types.
Use of circulating histone modifications in cancer patients
While the total amount of circulating nucleosomes is not cancer-specific, PTHMs in circulating nucleosomes could mirror cell-specific and disease-related processes and therefore could be a valuable source for novel biomarkers in cancer diagnostics. Initial studies of our group on this subject employed ChIP-based qPCR assays to detect and quantify PTHMs in blood circulation. In a first report, H3K9me3 and H4K20me3, hallmarks of pericentric heterochromatin (53), were investigated in blood plasma of patients with CRC, multiple myeloma and healthy controls using pericentric heterochromatin specific satellite 2 (29). H3K9me3 levels were found to be significantly decreased in patients with CRC whereas the decline for H4K20me3 was statistically not significant. The same approach was employed on a second set of cancer samples including CRC, breast and lung cancer as well as respective benign diseases and healthy individuals as controls. Both marks were found to be significantly decreased in CRC samples while increased in sera of breast cancer patients (30). In a subsequent study, deep sequencing of H4K20m3- and H3K9me3-related immunoprecipitated nucleosomal DNA was performed confirming the decrease of both markers in plasma of CRC patients as compared with healthy controls (54). Thereby, line-1 was found to be an abundant genetic marker that would be useful for further research. Given potential of these markers, we investigated H4K20me3 and H3K9me3 and another suppressive mark, H3K27me3, in CRC patients using an ELISA-based assay. Supporting previous results, we found reduced levels of H4K20me3 and H3K27me3 in CRC patients, in comparison to colonoscopy-verified cancer-free controls. However, unlike previous reports the levels of H3K9me3 were similar between the two groups. Encouragingly, when H3K27me3 and H4K20me3 were combined the results showed greater area under the curve (0.769), and sensitivity of 49.2% at 90% specificity for CRC (22). In another report with ELISA-based approach, it has also been demonstrated that H3K27 methylation levels can distinguish metastatic prostate cancer from organ confined, locally controlled disease (23).
Apart from our work, evidence for the clinical relevance of PTHMs in blood circulation is accumulating. An immunoassay for 5-methylated cytosines on circulating nucleosomes (5mc) reached 75% sensitivity at 70% specificity for detection of CRC versus healthy controls (55). In a Swedish study a panel of five epigenetic biomarkers (5mc, H2A.Z, H2A.A, H3K4me2 and H2AK119Ub) on circulating nucleosomes was superior to CA19-9 in patients with resectable pancreatic cancer in comparison to benign pancreatic disease and healthy controls (24). Combining CA19-9 with four of these epigenetic biomarkers increased the sensitivity in diagnosing pancreatic cancer. As an important precondition for retrospective analyses, the effects of pre-analytical variables were investigated recently to determine the stability of circulating nucleosomes under variable conditions. Thereby, parameters such as stasis, contamination with white cells, within-day variation, varying time before centrifugation, performance of colonoscopy and presence of a surgical trauma had no significant influence on the level of 5-methylcytosine DNA (5mc) or H3K9me3 in circulating nucleosomes (25). Furthermore, 5mc and H3K9me3 levels were significantly lower in cancer patients compared to healthy individuals. In a further study conducted in individuals referred to endoscopic screening for CRC, a combination of 12 different epigenetic marks were measured by ELISA assay and a panel of four markers provided an AUC of 0.97 enabling the discrimination of CRC from healthy controls with high sensitivity at early stages (sensitivity of 75 and 86 at 90% specificity for stages I and II, respectively) (26). Although numbers of investigated samples in these studies were limited, the findings confirm the potential of circulating nucleosome-associated epigenetic markers as a promising approach for cancer detection and differential diagnosis.
Conclusions and future perspectives
In recent years, “liquid biopsy” has gained considerable attention as a novel source of biomarkers and refers to the use of blood-based biomarkers in cancer detection and management. In addition to methylated circulating DNA and circulating non-coding RNAs, histone modifications in circulating nucleosomes generated a novel and promising class of epigenetic biomarkers in cancer. Several studies have provided proof-of-principle data on the potential of circulating histone marks such as methylation and acetylation for cancer detection. The combination of several histone marks rather than single histone marks could be utilized to enhance sensitivity and specificity of cancer detection. On the other side, different techniques including ChIP-PCR, ChIP-sequencing, ELISA-based assays or mass spectrometry were employed to detect and quantify PTHMs in serum or plasma. Sensitivity and specificity of PTHM detection using different techniques were not compared to each other. Furthermore, the reproducibility of results using different analytical methods across independent laboratories should be ensured before extensive, prospective trials are conducted to depict the clinical impact of PTHMs in cancer detection and diagnosis.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Heidi Schwarzenbach) for the series “Technologies in Liquid Biopsies - Potential applications in Medicine” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.09.42). The series “Technologies in Liquid Biopsies - Potential applications in Medicine” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Taby R, Issa JP. Cancer epigenetics. CA Cancer J Clin 2010;60:376-92. [Crossref] [PubMed]
- Kurdistani SK. Histone modifications in cancer biology and prognosis. Prog Drug Res 2011;67:91-106. [PubMed]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011;11:726-34. [Crossref] [PubMed]
- Audia JE, Campbell RM. Histone Modifications and Cancer. Cold Spring Harb Perspect Biol 2016;8:a019521 [Crossref] [PubMed]
- Gezer U, Holdenrieder S. Post-translational histone modifications in circulating nucleosomes as new biomarkers in colorectal cancer. In Vivo 2014;28:287-92. [PubMed]
- Yörüker EE, Holdenrieder S, Gezer U. Blood-based biomarkers for diagnosis, prognosis and treatment of colorectal cancer. Clin Chim Acta 2016;455:26-32. [Crossref] [PubMed]
- Hayes JJ, Hansen JC. Nucleosomes and the chromatin fiber. Curr Opin Genet Dev 2001;11:124-9. [Crossref] [PubMed]
- Mersfelder EL, Parthun MR. The tale beyond the tail: histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res 2006;34:2653-62. [Crossref] [PubMed]
- Cohen I, Poręba E, Kamieniarz K, et al. Histone modifiers in cancer: friends or foes? Genes Cancer 2011;2:631-47. [Crossref] [PubMed]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011;21:381-95. [Crossref] [PubMed]
- Li Y, Seto E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb Perspect Med 2016;6. [PubMed]
- Izzo A, Schneider R. Chatting histone modifications in mammals. Brief Funct Genomics 2010;9:429-43. [Crossref] [PubMed]
- Berry WL, Janknecht R. KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res 2013;73:2936-42. [Crossref] [PubMed]
- Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev 2007;21:3027-43. [Crossref] [PubMed]
- Jørgensen S, Schotta G, Sørensen CS. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res 2013;41:2797-806. [Crossref] [PubMed]
- Chervona Y, Costa M. Histone modifications and cancer: biomarkers of prognosis? Am J Cancer Res 2012;2:589-97. [PubMed]
- Tamagawa H, Oshima T, Shiozawa M, et al. The global histone modification pattern correlates with overall survival in metachronous liver metastasis of colorectal cancer. Oncol Rep 2012;27:637-42. [PubMed]
- Mosashvilli D, Kahl P, Mertens C, et al. Global histone acetylation levels: prognostic relevance in patients with renal cell carcinoma. Cancer Sci 2010;101:2664-9. [Crossref] [PubMed]
- Rothbart SB, Dickson BM, Raab JR, et al. An Interactive Database for the Assessment of Histone Antibody Specificity. Mol Cell 2015;59:502-11. [Crossref] [PubMed]
- Rossmann MP, Stillman B. Immunoblotting histones from yeast whole-cell protein extracts. Cold Spring Harb Protoc 2013;2013:625-30. [PubMed]
- Lakhal S, Mekki S, Ben-Abda I, et al. Evaluation of an enzyme-linked immunosorbent assay based on crude Leishmania histone proteins for serodiagnosis of human infantile visceral leishmaniasis. Clin Vaccine Immunol 2012;19:1487-91. [Crossref] [PubMed]
- Gezer U, Yörüker EE, Keskin M, et al. Histone Methylation Marks on Circulating Nucleosomes as Novel Blood-Based Biomarker in Colorectal Cancer. Int J Mol Sci 2015;16:29654-62. [Crossref] [PubMed]
- Deligezer U, Yaman F, Darendeliler E, et al. Post-treatment circulating plasma BMP6 mRNA and H3K27 methylation levels discriminate metastatic prostate cancer from localized disease. Clin Chim Acta 2010;411:1452-6. [Crossref] [PubMed]
- Bauden M, Pamart D, Ansari D, et al. Circulating nucleosomes as epigenetic biomarkers in pancreatic cancer. Clin Epigenetics 2015;7:106. [Crossref] [PubMed]
- Rasmussen L, Herzog M, Rømer E, et al. Pre-analytical variables of circulating cell-free nucleosomes containing 5-methylcytosine DNA or histone modification H3K9Me3. Scand J Clin Lab Invest 2016;76:448-53. [Crossref] [PubMed]
- Rahier JF, Druez A, Faugeras L, et al. Circulating nucleosomes as new blood-based biomarkers for detection of colorectal cancer. Clin Epigenetics 2017;9:53. [Crossref] [PubMed]
- Schones DE, Cui K, Cuddapah S. Genome-wide approaches to studying yeast chromatin modifications. Methods Mol Biol 2011;759:61-71. [Crossref] [PubMed]
- Deligezer U, Akisik EE, Erten N, et al. Sequence-specific histone methylation is detectable on circulating nucleosomes in plasma. Clin Chem 2008;54:1125-31. [Crossref] [PubMed]
- Deligezer U, Akisik EZ, Akisik EE, et al. H3K9me3/H4K20me3 ratio in circulating nucleosomes as potential biomarker for colorectal cancer. In: Gahan BP. editor. Circulating Nucleic Acids in Plasma and Serum. HK: Springer, 2009:97-103.
- Leszinski G, Gezer U, Siegele B, et al. Relevance of histone marks H3K9me3 and H4K20me3 in cancer. Anticancer Res 2012;32:2199-205. [PubMed]
- Freitas MA, Sklenar AR, Parthun MR. Application of mass spectrometry to the identification and quantification of histone post-translational modifications. J Cell Biochem 2004;92:691-700. [Crossref] [PubMed]
- Garcia BA, Shabanowitz J, Hunt DF. Characterization of histones and their post-translational modifications by mass spectrometry. Curr Opin Chem Biol 2007;11:66-73. [Crossref] [PubMed]
- Önder Ö, Sidoli S, Carroll M, et al. Progress in epigenetic histone modification analysis by mass spectrometry for clinical investigations. Expert Rev Proteomics 2015;12:499-517. [Crossref] [PubMed]
- Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 2005;37:391-400. [Crossref] [PubMed]
- Britton LM, Gonzales-Cope M, Zee BM, et al. Breaking the histone code with quantitative mass spectrometry. Expert Rev Proteomics 2011;8:631-43. [Crossref] [PubMed]
- Ziegler A, Zangemeister-Wittke U, Stahel RA. Circulating DNA: a new diagnostic gold mine? Cancer Treat Rev 2002;28:255-71. [Crossref] [PubMed]
- Jung M, Klotzek S, Lewandowski M, et al. Changes in concentration of DNA in serum and plasma during storage of blood samples. Clin Chem 2003;49:1028-9. [Crossref] [PubMed]
- Swinkels DW, Wiegerinck E, Steegers EA, et al. Effects of blood-processing protocols on cell-free DNA quantification in plasma. Clin Chem 2003;49:525-6. [Crossref] [PubMed]
- Holdenrieder S, Von Pawel J, Nagel D, et al. Long-term stability of circulating nucleosomes in serum. Anticancer Res 2010;30:1613-5. [PubMed]
- Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659-65. [PubMed]
- Jiang N, Reich CF 3rd, Pisetsky DS. Role of macrophages in the generation of circulating blood nucleosomes from dead and dying cells. Blood 2003;102:2243-50. [Crossref] [PubMed]
- Stroun M, Maurice P, Vasioukhin V, et al. The origin and mechanism of circulating DNA. Ann N Y Acad Sci 2000;906:161-8. [Crossref] [PubMed]
- Chan KC, Zhang J, Chan AT, et al. Molecular characterization of circulating EBV DNA in the plasma of nasopharyngeal carcinoma and lymphoma patients. Cancer Res 2003;63:2028-32. [PubMed]
- Snyder MW, Kircher M, Hill AJ, et al. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell 2016;164:57-68. [Crossref] [PubMed]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. [Crossref] [PubMed]
- Kuroi K, Tanaka C, Toi M. Plasma Nucleosome Levels in Node-Negative Breast Cancer Patients. Breast Cancer 1999;6:361-4. [Crossref] [PubMed]
- Holdenrieder S, Stieber P, Bodenmüller H, et al. Nucleosomes in serum of patients with benign and malignant diseases. Int J Cancer 2001;95:114-20. [Crossref] [PubMed]
- Stoetzer OJ, Fersching DM, Salat C, et al. Prediction of response to neoadjuvant chemotherapy in breast cancer patients by circulating apoptotic biomarkers nucleosomes, DNAse, cytokeratin-18 fragments and survivin. Cancer Lett 2013;336:140-8. [Crossref] [PubMed]
- Holdenrieder S, Stieber P, von Pawel J, et al. Circulating nucleosomes predict the response to chemotherapy in patients with advanced non-small cell lung cancer. Clin Cancer Res 2004;10:5981-7. [Crossref] [PubMed]
- Holdenrieder S, Stieber P. Therapy control in oncology by circulating nucleosomes. Ann N Y Acad Sci 2004;1022:211-6. [Crossref] [PubMed]
- Fahmueller YN, Nagel D, Hoffmann RT, et al. Predictive and prognostic value of circulating nucleosomes and serum biomarkers in patients with metastasized colorectal cancer undergoing Selective Internal Radiation Therapy. BMC Cancer 2012;12:5. [Crossref] [PubMed]
- Yörüker EE, Özgür E, Keskin M, et al. Assessment of circulating serum DNA integrity in colorectal cancer patients. Anticancer Res 2015;35:2435-40. [PubMed]
- Saksouk N, Simboeck E, Déjardin J. Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin 2015;8:3. [Crossref] [PubMed]
- Gezer U, Ustek D, Yörüker EE, et al. Characterization of H3K9me3- and H4K20me3-associated circulating nucleosomal DNA by high-throughput sequencing in colorectal cancer. Tumour Biol 2013;34:329-36. [Crossref] [PubMed]
- Holdenrieder S, Dharuman Y, Standop J, et al. Novel serum nucleosomics biomarkers for the detection of colorectal cancer. Anticancer Res 2014;34:2357-62. [PubMed]