
Exosomal lncRNAs as new players in cell-to-cell communication
Background
In order to survive, proliferate and metastasize cancer cells use various intercellular communication mechanisms (1). Classically the intercellular communication is classified as autocrine (local signaling between the same cell types), paracrine (local signaling between different cell types) and endocrine (distant signaling between any types of cells). Neoplastic cells use all these three communication categories in order to adapt to the local microenvironment, manipulate the immune system, and facilitate metastasis.
Non-coding RNA (ncRNAs) transcripts that do not codify proteins are widespread in the human genome and abnormally expressed in cancer cells (2). The most studied ncRNAs are the short microRNAs (miRNAs), produced from long primary ncRNAs such as miR-16-1/15a cluster from LEU2 transcript (3). Although the first ncRNA was discovered in bacteria in 1984 (4), only in the last decade careful investigation of these molecules in humans started.
An important class of ncRNAs is the long non-coding RNAs (lncRNAs). LncRNAs are transcripts over 200 nt long, frequently polyadenylated, alternatively spliced and with a 5’ terminal methylguanosine cap (5,6). LncRNAs exhibit little or no coding potential (7). Recently, the number of lncRNAs exceeded the number of mRNAs: to date, there are over 30,000 lncRNAs discovered and annotated (8) and this number will probably further increase. Initially considered “junk RNA”, lncRNA have just partially established roles in multiple organisms and cell types (9). Recent studies have revealed that lncRNA are crucial regulators of physiological processes (10,11) but are deregulated in various diseases, including cancer (12). These transcripts have a complex and thermodynamically stable secondary structure harboring hairpin loops, bulges, pseudoknots and double helices (13). The complex structure of the lncRNAs allows these molecules to interact with proteins, DNA, mRNA, and other non-coding classes of RNA (i.e., microRNA). LncRNAs play a complex role, coordinating the gene expression at multiple levels, from nuclear and cytoplasmic epigenetic processes (chromatin remodeling and microRNA sponging) to mRNA translation and decay (13).
In the last few years, lncRNAs were also identified in exosomes (14). Extracellular vesicles (EV) secretion is a new mechanism of cell-to-cell communication. Exosomes are the smallest subtype of EVs and also the most frequently associated with cancer (15). They are 30 to 100 nm in diameter nanovesicles, surrounded by a phospholipid bilayer. On the membrane surface, exosomes display several markers, including CD9, CD63, CD81, LAMP1 and TSG101 (16,17). Exosomes are generated by exocytosis of multivesicular bodies and are released into the interstitial spaces and into bodily fluids by normal and cancer cells (18,19). Secreted exosomes circulate in different body fluids and can be internalized by neighboring cells (autocrine and paracrine communication) or by remote recipient cells (endocrine communication) (20,21).
Exosomes enclose various types of molecules including lipids, proteins, DNA, mRNA and non-coding RNA (microRNA and lncRNA). Therefore, exosomes facilitate the exchange of genetic and epigenetic information between distant cells: this type of intercellular communication allows an epigenetic synchronization of cells from different tissues (22). Exosomes appear to be more than just vehicles of intercellular communication. Several studies described exosomes as a secretion mechanism discharging endogenous and exogenous waste products, toxic compounds, including chemotherapeutic drugs (23) and, probably, excessive intracellular lncRNAs.
The topic of cell-to-cell epigenetic communication via exosomal microRNAs is exhaustively summarized in several review papers (24-27). Therefore, in this article, we will review the role of exosomal lncRNAs in cancer, which is an incipient research field that could bring new insights to the vast domain of intercellular communication.
Exosome lncRNAs as potential cancer biomarkers
Liquid biopsy is a very attractive solution for a yet unsolved medical need – the early detection and treatment of neoplasia. The value of circulating lncRNA in predicting cancer is an intensely researched topic (28), but just a few papers assess if the lncRNAs are traveling via exosomes. There are three advantages in using exosomal lncRNAs as biomarkers: (I) lncRNAs inside the exosome are protected from RNases and can be presumed that the integrity and function of the lncRNAs are not altered (29); (II) the quantity of exosomal lncRNAs is high compared to other types of EVs (30); and (III) the expression of many lncRNAs is tissue specific (31). Therefore, profiling the exosomal lncRNAs can lead to the development of valuable diagnostic markers.
Several papers propose exosomal lncRNAs as sensible and specific new biomarkers for different types of cancer. An RNA panel, including 21 lncRNAs, has been found significantly different in exosomes of colorectal cancer (CRC) patients. The combination of two exosomal mRNAs and one lncRNA—breast cancer anti-estrogen resistance 4 (BCAR4), showed a good cancer prediction efficiency, with an AUC of 0.877. Furthermore, exosomal BCAR4 was also found decreased in the serum of patients with colon adenoma compared to healthy subjects, suggesting its low level plays a role in the CRC development (30). An increased level of tumor released lncRNA, colorectal neoplasia differentially expressed-h (CRNDE-h) was found in the exosomes of CRC patients proving its efficiency in identifying these neoplastic patients. The screening efficiency to predict CRC increases by combining exosomal CRNDE-h and CEA, in comparison to using a single biomarker, reaching an AUC of 0.913. A high exosomal CRNDE-h was positively correlated with the patients’ metastatic status and proved to be an independent prognostic factor for overall survival in a cohort of 148 patients (32).
Exosomal long intergenic non-protein-coding RNA 152 (LINC00152) is an lncRNA reported as overexpressed in plasma of gastric cancer (GC) patients. Because there is no difference between exosomal LINC00152 levels and total LINC00152 plasma levels, researchers supposed the plasma LINC00152 was traveling exclusively via exosomes. After applying the ROC curve the authors determine that exosomal LINC00152 is a more sensitive marker compared to the established, CEA and CA199 (33). Zinc finger antisense 1 (ZFAS1), is a serum circulating exosomal lncRNA which is also upregulated in GC, the estimated specificity and sensibility for the diagnosis of GC of ZFAS1 is 80.0% and 75.7%, respectively. The authors further demonstrated that GC cell lines can internalize exosomes containing ZFAS1 resulting in a more aggressive phenotype (34).
Zhang et al. identified the exosomal, lncRNA, Metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1) to be overexpressed in the serum of non-small cell lung cancer (NSCLC) patients. The authors tested the potential role of MALAT-1 and discovered a sensitivity and specificity of 60.1% and 80.9%, respectively. Additionally, in vitro studies demonstrated that the overexpression of MALAT-1 promotes migration and cell proliferation (35).
Hox antisense intergenic RNA (HOTAIR) is one of the most frequently reported lncRNAs involved in cancer development. Wang et al. found that the exosomal HOTAIR could be a diagnosis marker of laryngeal squamous cell carcinoma (LSCC). The screening sensitivity and specificity of HOTAIR increases when combined with miR-21. High levels of exosomal HOTAIR also correlate with the clinical stage of LSCC (36).
Besides serum exosome lncRNAs, there are some studies focusing on the role of exosomal lncRNAs contained in other bodily fluids. Urine exosomes containing lncRNAs are regarded as potential biomarkers for urothelial bladder cancer (UBC) and prostate cancer. HOTAIR is overexpressed in urine exosomes of bladder cancer patients and correlates with the local invasion status of the tumor. The in vitro knock-down of HOTAIR leads to a decrease in migration, a less invasive phenotype and reduced epithelial to mesenchymal transition (EMT) factors expression. The same research group reported seven more lncRNAs that are overexpressed in urine exosomes of invasive UBC patients: HOXA cluster antisense RNA 2 (HOX-AS-2), MALAT-1, Long intergenic non-protein coding RNA regulator of reprogramming (LINC-ROR), Hydatidiform mole associated and imprinted non-protein coding RNA 1 (HYMA1), Long intergenic non-protein coding RNA 477 (LINC00477), LOC 100506688, and Orthodenticle homeobox 2 antisense RNA 1 (OTX2-AS1) (29). The lncRNA, Prostate cancer antigen 3 (PCA3), is an already FDA approved urine biomarker for prostate adenocarcinoma. Most studies do not characterize the way PCA3 travels in urine. Recently, it was found that PCA3 accumulates in urine exosomes and does not travel free in urine. The pressure exerted on the prostate gland by digital rectal examination leads to an increased level of exosomal PCA3 in urine facilitating the prostate cancer diagnosis (37). In addition, it was discovered that urine exosomal lncRNA-p21 is suitable to differentiate prostate adenocarcinoma from benign prostate hyperplasia (38).
Another study showed that the cervicovaginal lavage from cervical cancer patients is enriched in exosomes containing the lncRNAs, HOTAIR, MALAT-1 and maternally expressed 3 (MEG-3). Of these exosomal lncRNAs, HOTAIR and MALAT-1 are overexpressed in cervical cancer patients. In contrast, MEG-3 is down regulated in cancer patient lavage compared to the control group. Further analysis revealed that HPV infection might regulate the exosomal lncRNAs secretion, since the level of these transcripts varies between HPV positive patients and the negative control group (39).
All the data regarding exosomal lncRNAs as potential diagnosis and prognostic biomarkers are summarized in Table 1.
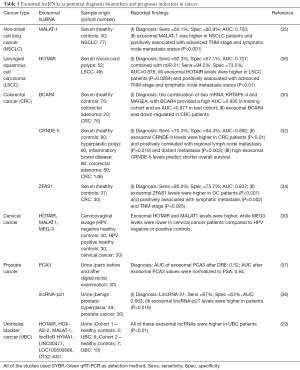
Full table
Roles of exosomal lncRNAs
The role of exosomal lncRNAs is not completely characterized. Recent studies show that exosomal lncRNAs can travel to neighboring and distant cells where they transmit phenotypical changes such as drug resistance, promote angiogenesis and induce tumorigenesis (Figure 1).
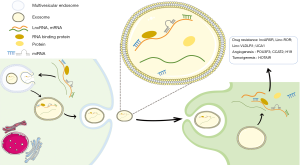
Drug resistance
Qu et al. discovered a new lncRNA, lncRNA Activated in RCC with Sunitinib Resistance (lncARSR), promoting sunitinib resistance and predicting poor therapeutic response in renal cell carcinoma patients (40). They revealed, in vitro and in vivo, that the sunitinib resistance disseminates between cancer cells via exosomes containing lncARSR. Additionally, they showed that lncARSR acts as a competitive-endogenous RNA (ceRNA), that sponges miR-34 and miR-449 in order to promote the expression of AXL and c-MET, oncogenes that are responsible for the drug resistance (40).
LINC-ROR is an lncRNA that plays a role in TGFβ mediated chemoresistance and respond to hypoxia in hepatocellular carcinoma (HCC) (41). In vitro exposure of HCC cells to TGFβ (a protein already associated with chemotherapy resistance) or direct exposure to sorafenib, camptothecin and doxorubicin induces an increase in exosomes loaded with LINC-ROR. LINC-ROR knockdown in HCC cells determines early apoptosis, increase of caspase 3/7 activity, and reduces the cell survival. This combined data could suggest that LINC-ROR is a modulator of chemotherapy resistance in HCC (42). The same research group discovered a second lncRNA involved in HCC chemotherapy resistance. LincRNA-VLDLR (linc-VLDLR) is overexpressed in HCC cell lines and in the exosomes derived from these cells after exposure to sorafenib, camptothecin and doxorubicin. The mechanism by which linc-VLDLR contributes to anticancer drug resistance is not yet clear, but it seems that the incubation of HCC cells with exosomes derived from resistant cells leads to overexpression of ABC subfamily G member 2, a protein involved in extracellular drug export (43).
Xu et al. showed that exosomes from tamoxifen resistant breast cancer cells contain significantly higher levels of the lncRNA—urothelial cancer associated 1 (UCA1) compared to tamoxifen sensitive cells. Furthermore, the tamoxifen resistant phenotype can be transferred through exosomes containing UCA1 (44).
Angiogenesis
The lncRNA POU class 3 homeobox 3 (POU3F3) is overexpressed in glioma tissue compared to the adjacent normal tissue. In order to demonstrate the function of this lncRNA, exosomes from A172 glioma cell lines expressing a high level of PUO3F3 were isolated and co-cultured with human brain microvascular endothelial cells (HBMECs). Using immunofluorescence staining it was observed that HBMECs internalize the exosomes containing high levels of PUO3F3. The internalization of the exosomes leads to an increase in cell proliferation, migration, tube formation, and in vivo angiogenesis. They further confirmed the angiogenetic function of POU3F3 in HBMECs by detecting an increased level of proteins involved in neo-vascularization (45).
Using similar methods, the same research group demonstrated that the exosomal Colon cancer associated transcript 2 (CCAT2), an lncRNA cloned at 8q24 amplicon (46,47), promotes cell proliferation, migration, tube formation in vitro and angiogenesis in vivo. Additionally, the authors demonstrate that CCAT2 derived from glioma cells can inhibit the apoptosis of endothelial cells (HUVECs) (48).
H19 is an lncRNA up-regulated in the exosomes of CD90+ HCC cells and has angiogenetic potential. The incubation of HUVECs with exosomes derived from CD90+ cells stimulates the increase of intercellular adhesion proteins and of VEGF and VEGF-R. Similar results were obtained also after inducing H19 overexpression in HUVECs (49). In conclusion, all these studies show that tumor cells can change the phenotype of endothelial cells through lncRNAs contained into exosomes.
Mediators of cancer risk factors
In a very interesting paper, Lu et al. explored the mechanism that links sedentary life style and obesity to the risk of developing CRC (50). They discovered in a mouse model that the circulating exosomal HOTAIR is secreted by the gluteal-femoral fat and travels to intestinal stem cells where the exosomes are internalized. Additionally, in vitro studies unveiled that exosomal HOTAIR is an activator of the Wnt pathway of intestinal cells. Their clinical studies confirmed the overexpression of exosomal HOTAIR in obese patients with sedentary life-style (50). Further clinical studies are required to prove that increased HOTAIR levels correlate with the risk of developing CRC.
Mechanistic insights regarding exosomal lncRNAs
One can presume that lncRNAs and other RNA species appear to be loaded selectively into exosomes and represent new shuttles of cell-to-cell communication. The expression level of exosomal lncRNAs varies from its donor cells (51-54). The level of the oncogenic lncRNA TUC339 is dramatically different in EV, including exosomes, compared to HCC donor cells (53). It was discovered that the expression level of seven lncRNAs differs between the cultured cancer cell lines and exosomes (51). These experiments clearly suggest that lncRNAs are not randomly secreted in the extracellular milieu via exosomes. Moreover, using RNA-Seq, Chen et al., confirmed that exosomes from LIM1863, a polarized colon carcinoma cell line with cryptic-like structure, are preferentially enriched in 2,389 mRNAs, 317 pseudogenes, 1,028 lncRNAs, and 206 short ncRNAs compared to parental LIM1863 whole cell lysates. They further classified the exosomes in apical and basolateral type and demonstrated the RNA signature differs between the two subclasses (54).
The mechanism by which lncRNAs are loaded into exosomes is not yet understood, and has to be deciphered in parallel with the mechanism by which microRNAs and other small RNAs are internalized into exosomes. LncRNAs most probably contain RNA structure motifs and protein partners that are responsible for the intracellular localization (55) and exosomal loading. Additionally, because of the complex structure of lncRNAs, they can also be scaffolders for other molecules (i.e., miRNAs, mRNAs, and proteins) and facilitate their exosomal loading.
It was showed that Promoter of MAT2A-antisense radiation induced circulating lncRNA, (PARTICLE), is responsible for the fine tuning of MAT2A-SAM-SAH-homocysteine pathway in response to irradiation (56). PARTICLE exerts its function in the nucleus interacting directly with the DNA by controlling the methylation of the MAT2A promoter region, and in the cytoplasm where it binds with MAT2A transcripts. Furthermore, the authors showed that PARTICLE and MAT2A levels are simultaneously overexpressed in exosomes after irradiation. The incubation of naïve cells with these exosomes enriched in PARTICLE and MAT2A leads to overexpression of intracellular PARTICLE. Increased levels of PARTICLE were also detected in plasma samples of post-radiation therapy patients. The authors proposed PARTICLE and MAT2A as potential biomarkers in patient post-radiotherapy and hypothesized that PARTICLE acts as a loading vehicle for MAT2A into exosomes (56).
It has been hypothesized that lncRNAs, pseudogenes and mRNAs crosstalk by competing for a common pool of miRNAs and build a complex regulatory network. This theory is called the competitive endogenous RNA (ceRNA) hypothesis (57). Recently, multiple researchers used mathematical and experimental models to contest the ceRNA hypothesis (58,59). The counter-argument of these researchers is that miRNA binding sites harbored by mRNA and lncRNA never reach high enough levels in order for the RNA regulatory crosstalk to exist. Using quantitative computational models, they estimate that only high, unphysiological levels of binding sites could make the miRNA sponging possible (58,59). The ceRNA crosstalk is highly dependent on the subcellular localization of the involved RNAs, where higher concentrations of the interacting partners are reached (60) and therefore ceRNA could be dependent of cellular/extracellular compartmentalization. One can presume that exosomes can be the ideal place for RNA species interaction where unphysiological concentrations of miRNA binding sites are reached and crosstalk may occur. The experiments performed by Ahadi et al., partially sustain this hypothesis (61). Using lncRNA arrays, the authors discover that 26 lncRNAs are overexpressed in exosomes from prostate cancer cell lines compared to normal epithelial. In order to understand the role of these lncRNAs, they predicted microRNAs seed regions of these lncRNAs and confirmed the high expression level of the predicted targets in prostate cancer cell lines exosomes (61).
Final remarks
lncRNAs biomarkers
Exosomal lncRNAs seem to be promising cancer biomarkers. Most of the studies that research lncRNAs as diagnostic tools do not show if these transcripts are located into exosomes. We consider that assessing the mechanism by which lncRNAs travel in bodily fluids is important. This step opens up new research pathways helping us understand the roles of these molecules and understand their diagnostic potential. Studying the membrane proteins of the exosome can give hints about the origin and the destination of the lncRNAs (62). Moreover, one can also speculate that exosomal lncRNAs are functional and integral when compared to lncRNAs that travel free, as exosomal lncRNAs are protected from the ubiquitary presence of RNase. Future studies, using higher patient numbers are needed to confirm the role of lncRNAs as cancer biomarkers. Additionally, it is very important to standardize preanalytic and analytic procedures in order to reach high reproducibility. However, one can ask himself how disease specific are these lncRNAs. We can simply observe that the same lncRNAs are proposed as highly specific biomarkers for several cancers as well as various non-neoplastic diseases. For example, the already famous HOTAIR and MALAT1 are proposed as biomarkers for different types of cancer. The same phenomenon has been observed in the case of microRNAs (63) and circulating proteins (64). One possible solution to overcome this issue of low specificity is to analyze more than one circulating molecule and build networks with direct and indirect interactions between the molecules (65,66). Another solution is to integrate the membrane proteins of exosomes in the diagnostic algorithm. Future studies are needed to prove if the membrane structure analysis permits the detection of the tissue of origin of exosomes.
The role of exosomal lncRNAs
Numerous studies show that cancer derived exosomal lncRNAs are functional and can transmit to neighboring cells different phenotypic patterns, like drug resistance and increased angiogenesis. We remark, that most of the studies focus on exosomal lncRNAs that are secreted by the cancer cell and are incorporated in neighboring cancer cells or in the microenvironment. Only one study explores the exosomal lncRNAs that are secreted by nonmalignant tissues and are internalized by premalignant cells. The authors show that exosomal HOTAIR is secreted by adipose tissue and internalized by intestinal cells which start proliferating. This observation links sedentary life style and obesity to the development of CRC (50). Exosomal cell-to-cell communications can help us understand epidemiological correlations otherwise unexplainable. Communication by exosomal lncRNAs can also shed light on the complex interaction between the immune system and the tumor tissue, an area unexplored from this point of view. Additionally, there are no in vivo studies that investigate the role of exosomal lncRNAs in the development of distant metastasis.
Mechanistic insights regarding exosomal lncRNAs function. Most studies asses the expression levels of only one exosomal lncRNA. We consider it is important to describe the level of several lncRNAs and other RNA species that travel via exosomes and to characterize their interaction. The exosome can be also perceived as a cocktail of molecules that reach unphysiological concentration that could co-interact. Hence, the exosome can be a suitable study model for the ceRNA hypothesis. Finally, we observe that just a few studies research the loading mechanism of lncRNAs into exosomes. LncRNAs could be loading vehicles for miRNAs, mRNAs and other small molecules into the exosome. Future studies are required to confirm this supposition.
Exosomal lncRNAs as therapeutic targets
Manipulating the exosomal lncRNAs is also an attractive therapeutic method because exosomes have an excellent biocompatibility and biodistribution compared to other drug delivery systems (67). We can imagine two possible strategies to use exosomal lncRNAs as therapeutic targets. Firstly, exosomes can be used as a delivery vehicle for lncRNAs with tumor suppressor function. In order to do this, it is crucial to determine which factors control the internalization of exosomes in a specific cell type or tissue. Also, attention must be taken to avoid any unwanted immune reaction generated by exosomes. Secondly, in the case of lncRNAs which stimulate neo-vascularization and transmit drug resistance, we can imagine a therapeutic strategy that interrupts an exosomal lncRNA intercellular communication pathway. Understanding the mechanisms by which lncRNAs are loaded into exosomes, could lead to the development of this new therapeutic method that blocks the internalization of lncRNAs into exosomes.
Although important steps have been made in understanding this new pathway of cell-to-cell communication, it is clear that further preclinical investigations are needed to move this interesting research domain from bench to bedside.
Acknowledgments
We are thankful to Mr. Sergiu Dragomir and Mr. Timothy Keegan for critical reading the manuscript. The authors would also like to thank Servier Medical Art used to create the figure.
Funding: Work in Dr. Calin’s laboratory is supported by National Institutes of Health (NIH/NCATS) grant UH3TR00943-01 through the NIH Common Fund, Office of Strategic Coordination (OSC), the NIH/NCI grant 1 R01 CA182905-01, a U54 grant—UPR/MDACC Partnership for Excellence in Cancer Research 2016 Pilot Project, a Team DOD (CA160445P1) grant, a Ladies Leukemia League grant, a CLL Moonshot Flagship project, a SINF 2017 grant, and the Estate of C. G. Johnson, Jr. The work of M Dragomir is supported by a POC grant, entitled “Clinical and economical impact of personalized targeted anti-microRNA therapies in reconverting lung cancer chemoresistance”—CANTEMIR, Competitivity Operational Program, 2014-2020, no. 35/01.09.2016, MySMIS 103375.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Heidi Schwarzenbach) for the series “Technologies in Liquid Biopsies - Potential applications in Medicine” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ tcr.2017.10.46). The series “Technologies in Liquid Biopsies - Potential applications in Medicine” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0.
References
- Brucher BL, Jamall IS. Cell-cell communication in the tumor microenvironment, carcinogenesis, and anticancer treatment. Cell Physiol Biochem 2014;34:213-43. [Crossref] [PubMed]
- Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 2002;99:15524-9. [Crossref] [PubMed]
- Bullrich F, Fujii H, Calin G, et al. Characterization of the 13q14 tumor suppressor locus in CLL: identification of ALT1, an alternative splice variant of the LEU2 gene. Cancer Res 2001;61:6640-8. [PubMed]
- Jarroux J, Morillon A, Pinskaya M. History, Discovery, and Classification of lncRNAs. Adv Exp Med Biol 2017;1008:1-46. [Crossref] [PubMed]
- Ulitsky I, Bartel DP. lincRNAs: Genomics, Evolution, and Mechanisms. Cell 2013;154:26-46. [Crossref] [PubMed]
- Spizzo R, Almeida MI, Colombatti A, et al. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene 2012;31:4577-87. [Crossref] [PubMed]
- Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775-89. [Crossref] [PubMed]
- Flicek P, Amode MR, Barrell D, et al. Ensembl 2014. Nucleic Acids Res 2014;42:D749-55. [Crossref] [PubMed]
- Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol 2015;22:5-7. [Crossref] [PubMed]
- Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 2014;14:752-61. [Crossref] [PubMed]
- Grote P, Wittler L, Hendrix D, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 2013;24:206-14. [Crossref] [PubMed]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol 2011;21:354-61. [Crossref] [PubMed]
- Mercer TR, Mattick JS. Understanding the regulatory and transcriptional complexity of the genome through structure. Genome Res 2013;23:1081-8. [Crossref] [PubMed]
- Huang X, Yuan T, Tschannen M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 2013;14:319. [Crossref] [PubMed]
- Zhang HG, Grizzle WE. Exosomes: a novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am J Pathol 2014;184:28-41. [Crossref] [PubMed]
- Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 2010;73:1907-20. [Crossref] [PubMed]
- Dignat-George F, Boulanger CM. The Many Faces of Endothelial Microparticles. Arterioscler Thromb Vasc Biol 2011;31:27-33. [Crossref] [PubMed]
- Gyorgy B, Szabo TG, Pasztoi M, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci 2011;68:2667-88. [Crossref] [PubMed]
- Kim KM, Abdelmohsen K, Mustapic M, et al. RNA in extracellular vesicles. Wiley Interdiscip Rev RNA 2017;8. [PubMed]
- Melo SA, Sugimoto H, O'Connell JT, et al. Cancer Exosomes Perform Cell-Independent MicroRNA Biogenesis and Promote Tumorigenesis. Cancer Cell 2014;26:707-21. [Crossref] [PubMed]
- Peinado H, Alečković M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012;18:883-91. [Crossref] [PubMed]
- Mittelbrunn M, Sanchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol 2012;13:328-35. [Crossref] [PubMed]
- Shedden K, Xie XT, Chandaroy P, et al. Expulsion of small molecules in vesicles shed by cancer cells: Association with gene expression and chemosensitivity profiles. Cancer Res 2003;63:4331-7. [PubMed]
- Braicu C, Tomuleasa C, Monroig P, et al. Exosomes as divine messengers: are they the Hermes of modern molecular oncology? Cell Death Differ 2015;22:34-45. [Crossref] [PubMed]
- Sato-Kuwabara Y, Melo SA, Soares FA, et al. The fusion of two worlds: Non-coding RNAs and extracellular vesicles - diagnostic and therapeutic implications. Int J Oncol 2015;46:17-27. [Crossref] [PubMed]
- Tovar-Camargo OA, Toden S, Goel A. Exosomal microRNA Biomarkers: Emerging Frontiers in Colorectal and Other Human Cancers. Expert Rev Mol Diagn 2016;16:553-67. [Crossref] [PubMed]
- Schwarzenbach H. The clinical relevance of circulating, exosomal miRNAs as biomarkers for cancer. Expert Rev Mol Diagn 2015;15:1159-69. [Crossref] [PubMed]
- Huarte M. The emerging role of IncRNAs in cancer. Nat Med 2015;21:1253-61. [Crossref] [PubMed]
- Berrondo C, Flax J, Kucherov V, et al. Expression of the Long Non-Coding RNA HOTAIR Correlates with Disease Progression in Bladder Cancer and Is Contained in Bladder Cancer Patient Urinary Exosomes. PLoS One 2016;11:e0147236 [Crossref] [PubMed]
- Dong L, Lin WR, Qi P, et al. Circulating Long RNAs in Serum Extracellular Vesicles: Their Characterization and Potential Application as Biomarkers for Diagnosis of Colorectal Cancer. Cancer Epidemiol Biomarkers Prev 2016;25:1158-66. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- Liu T, Zhang X, Gao S, et al. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget 2016;7:85551-63. [Crossref] [PubMed]
- Li Q, Shao Y, Zhang X, et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol 2015;36:2007-12. [Crossref] [PubMed]
- Pan L, Liang W, Fu M, et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol 2017;143:991-1004. [Crossref] [PubMed]
- Zhang R, Xia Y, Wang Z, et al. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun 2017;490:406-14. [Crossref] [PubMed]
- Wang J, Zhou Y, Lu J, et al. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med Oncol 2014;31:148. [Crossref] [PubMed]
- Dijkstra S, Birker IL, Smit FP, et al. Prostate cancer biomarker profiles in urinary sediments and exosomes. J Urol 2014;191:1132-8. [Crossref] [PubMed]
- Isin M, Uysaler E, Ozgur E, et al. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front Genet 2015;6:168. [PubMed]
- Zhang J, Liu SC, Luo XH, et al. Exosomal Long Noncoding RNAs are Differentially Expressed in the Cervicovaginal Lavage Samples of Cervical Cancer Patients. J Clin Lab Anal 2016;30:1116-21. [Crossref] [PubMed]
- Qu L, Ding J, Chen C, et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell 2016;29:653-68. [Crossref] [PubMed]
- Takahashi K, Yan IK, Haga H, et al. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J Cell Sci 2014;127:1585-94. [Crossref] [PubMed]
- Takahashi K, Yan IK, Kogure T, et al. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio 2014;4:458-67. [Crossref] [PubMed]
- Takahashi K, Yan IK, Wood J, et al. Involvement of extracellular vesicle long noncoding RNA (linc-VLDLR) in tumor cell responses to chemotherapy. Mol Cancer Res 2014;12:1377-87. [Crossref] [PubMed]
- Xu CG, Yang MF, Ren YQ, et al. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur Rev Med Pharmacol Sci 2016;20:4362-8. [PubMed]
- Lang HL, Hu GW, Chen Y, et al. Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur Rev Med Pharmacol Sci 2017;21:959-72. [PubMed]
- Ling H, Spizzo R, Atlasi Y, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res 2013;23:1446-61. [Crossref] [PubMed]
- Redis RS, Vela LE, Lu W, et al. Allele-Specific Reprogramming of Cancer Metabolism by the Long Non-coding RNA CCAT2. Mol Cell 2016;61:520-34. [Crossref] [PubMed]
- Lang HL, Hu GW, Zhang B, et al. Glioma cells enhance angiogenesis and inhibit endothelial cell apoptosis through the release of exosomes that contain long non-coding RNA CCAT2. Oncol Rep 2017;38:785-98. [Crossref] [PubMed]
- Conigliaro A, Costa V, Lo Dico A, et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer 2015;14:155. [Crossref] [PubMed]
- Lu X, Bai D, Liu X, et al. Sedentary lifestyle related exosomal release of Hotair from gluteal-femoral fat promotes intestinal cell proliferation. Sci Rep 2017;7:45648. [Crossref] [PubMed]
- Gezer U, Ozgur E, Cetinkaya M, et al. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int 2014;38:1076-9. [PubMed]
- Koldemir O, Ozgur E, Gezer U. Accumulation of GAS5 in exosomes is a marker of apoptosis induction. Biomed Rep 2017;6:358-62. [Crossref] [PubMed]
- Kogure T, Yan IK, Lin WL, et al. Extracellular Vesicle-Mediated Transfer of a Novel Long Noncoding RNA TUC339: A Mechanism of Intercellular Signaling in Human Hepatocellular Cancer. Genes Cancer 2013;4:261-72. [Crossref] [PubMed]
- Chen M, Xu R, Ji H, et al. Transcriptome and long noncoding RNA sequencing of three extracellular vesicle subtypes released from the human colon cancer LIM1863 cell line. Sci Rep 2016;6:38397. [Crossref] [PubMed]
- Chen LL. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci 2016;41:761-72. [Crossref] [PubMed]
- O'Leary VB, Ovsepian SV, Carrascosa LG, et al. PARTICLE, a Triplex-Forming Long ncRNA, Regulates Locus-Specific Methylation in Response to Low-Dose Irradiation. Cell Rep 2015;11:474-85. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Jens M, Rajewsky N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat Rev Genet 2015;16:113-26. [Crossref] [PubMed]
- Denzler R, Agarwal V, Stefano J, et al. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell 2014;54:766-76. [Crossref] [PubMed]
- Vasilescu C, Tanase M, Dragomir M, et al. From mobility to crosstalk. A model of intracellular miRNAs motion may explain the RNAs interaction mechanism on the basis of target subcellular localization. Math Biosci 2016;280:50-61. [Crossref] [PubMed]
- Ahadi A, Brennan S, Kennedy PJ, et al. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci Rep 2016;6:24922. [Crossref] [PubMed]
- Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem 2015;61:56-63. [Crossref] [PubMed]
- Haider BA, Baras AS, McCall MN, et al. A critical evaluation of microRNA biomarkers in non-neoplastic disease. PLoS One 2014;9:e89565 [Crossref] [PubMed]
- Maestranzi S, Przemioslo R, Mitchell H, et al. The effect of benign and malignant liver disease on the tumour markers CA19-9 and CEA. Ann Clin Biochem 1998;35:99-103. [Crossref] [PubMed]
- Volinia S, Galasso M, Costinean S, et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res 2010;20:589-99. [Crossref] [PubMed]
- Vasilescu C, Dragomir M, Tanase M, et al. Circulating miRNAs in sepsis-A network under attack: An in-silico prediction of the potential existence of miRNA sponges in sepsis. PLoS One 2017;12:e0183334 [Crossref] [PubMed]
- Kamerkar S, LeBleu VS, Sugimoto H, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017;546:498-503. [PubMed]


