
Untargeted profiling of cell-free circulating DNA
Introduction
Cell-free circulating tumor DNA (ctDNA) is released from tumors and reflects their genomic landscapes, making it a perfect minimally invasive biomarker. In recent years, it is has been shown in numerous studies that the analysis of ctDNA is a very powerful tool and might revolutionize cancer care with respect to early detection, identification of minimal residual disease, assessment of treatment response, and monitoring tumor evolution (1-5). For example, a variety of studies have found decreasing levels of ctDNA after surgery and/or chemotherapy and have shown that changing levels of ctDNA can be used to assess response to treatment (6-8). In a landmark study, Dawson et al. were able to demonstrate in metastatic breast cancer (MBC) patients that ctDNA levels showed a greater dynamic range and greater correlation with changes in tumor burden than the conventional tumor marker CA15.3 or circulating tumor cells (CTCs) (6). In a follow-up study, the first exome-wide sequencing analysis of tumor cfDNA was performed and the authors were able to track acquired resistance to cancer therapy in almost all patients (9). However, several studies have demonstrated that, despite the assumption that ctDNA levels correlate with tumor burden, occasionally patients with large tumors do not have detectable amounts of ctDNA in their circulation and the reasons for these variable results remain unknown (10-12). In a study employing low coverage whole genome sequencing (WGS) from our group, we showed that in some breast cancer patients, unexpectedly low fractions of ctDNA, despite metastatic disease, can be observed, suggesting that the ctDNA levels reflect the progression/proliferation status of a tumor rather than the actual tumor burden (10).
The recognition of the clinical utility of ctDNA analyses has led to technical improvements, now enabling the analysis of single mutations with a high resolution (13,14). In this regard, most studies employing ctDNA rely on the interrogation of a small set of genes or hotspot mutations. However, due to the fact that tumor genomes are constantly changing, a comprehensive genome-wide analysis for capturing novel occurring alterations under the selective pressure of targeted therapies has proven to be beneficial, at least in late-stage cancer patients. Here, we review studies which applied untargeted approaches for ctDNA analyses, highlight the advantages over targeted approaches and discuss related limitations.
Increasing genomic coverage comes with decreasing analytical sensitivity
Due to technical improvements, a variety of methods such as digital PCR or in-depth resequencing can reach a high resolution and are able to detect a few mutant alleles in the background of thousands of wild type alleles. It is of note, though, that a sensitivity of 0.01% and lower can only be achieved with sufficient input amounts of DNA fragments. With respect to conventional next generation sequencing (NGS)-based approaches, a reliable detection of underrepresented alleles (<1–5%) cannot be assured, as analytical sensitivity is dependent on several factors. Already during library preparation, PCR artefacts can be introduced as a result of polymerases errors, low input amounts or a high number of PCR cycles. Moreover, the sequencing strategy has to be considered, as different sequencing platforms have different random error rates ranging from 0.1% to 1%. Furthermore, the accuracy of variant calling depends on the depth of sequence coverage, thus the identification of a variant improves with increasing coverage. Finally, base calling can be influenced by local sequence context and therefore the assessment of general sensitivity may not always be true for every possible variant (15). Due to high rates of false positives with traditional NGS, most methods interrogate only a few targets or employ a gene panel in cases where the ctDNA fraction is greater than 1-5% of total circulating cfDNA. However, a variety of genomic alterations may be missed if the analysis is only limited to hotspots. Therefore, more comprehensive and efficient strategies with high resolution are needed in order to identify all actionable genomic alterations within a sample (16). The development of molecular barcoding approaches has greatly improved the analytical sensitivity of NGS approaches (17,18). In recent years, efforts have been made to develop targeted approaches which are able to interrogate up to one hundred genes instead of single genes by employing molecular barcoding approaches. These methods are either based on amplicon sequencing or utilize hybridization-based capturing [e.g., CAPP-Seq (19,20)]. In addition, during the last 2 years, several companies have launched so-called hotspot panels for the analysis of up to 200 common cancer targets from 20–50 genes, which are applicable for cfDNA (e.g., AmpliSeq Cancer panel, Life Tech; TruSeq Cancer Amplicon, Illumina; the Human Actionable Solid Tumor Panel, Qiagen; NEBNext Direct® Cancer HotSpot Panel, NEB; AVENIO ctDNA Targeted Kit, Roche). Most of these companies also offer entity-specific or customizable panels enabling an individualized composition of target genes. The target variant allele frequencies (VAF) of these assays vary from 0.05% to >1% depending on the specific application of interest, the panel size, sample multiplexing, and the number of sequencing reads. Yet, data regarding the application of such panels are sparse and detailed assessments of sensitivity and specificity from independent research groups have not yet been published.
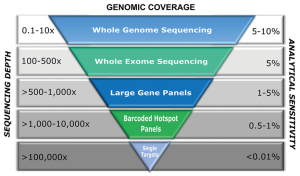
The influence on sequencing coverage depth on detection rate of variants was recently demonstrated from a group in China, who implemented a customized gene panel of 382 cancer-relevant genes on 605 ctDNA samples in multiple cancer types to assess the feasibility of comprehensive ctDNA mutation profiling to guide treatment decisions in cancer (21). Using a VAF cut-off of 1%, they were able to identify somatic mutations in 87% of ctDNA samples, with mutation spectra highly concordant with matched tumor tissues (21). Not surprisingly, VAFs in ctDNA were significantly lower and 66% of mutations demonstrated a VAF below 10% (median: 5%), while mutations in tumor tissues had a median VAF of 23%. When using <300X mean coverage depth of ctDNA, only 76% of patients showed mutations while a coverage depth of 300–500X yielded an 87% detection rate and concordance between tumor and ctDNA increased from 67% to 88%. However, by applying the 1% VAF threshold, both concordance and detection rate hardly improved when the coverage depth was increased up to 2,000X. Similarly, whole-exome sequencing from plasma has demonstrated high levels of concordance between mutations in plasma DNA of breast cancer patients with metastatic late stage disease and the respective tumor tissue (9). It must be pointed out that the samples analysed in this study had exceptionally high ctDNA levels ranging from 33–65%. Butler et al. were able to reliably recapitulate the tumor genome from plasma even in a sample with an average VAF of 3.7% (22). This VAF range is, however, currently the maximum achievable analytical sensitivity of exome sequencing. Although WES already enables a comprehensive view of tumor-derived genetic changes and can to some extent inform about rearrangements or copy number alterations, a global genomic profiling of ctDNA aberrations can only be achieved by WGS. However, since analytical sensitivity is, among other factors, dictated by sequencing depth, a comparable detection limit for mutations at the sequence level as with WES is prohibitively expensive for WGS (Table 1). Even for the detection of rearrangements at high resolution, sequence coverage of at least 20X at high ctDNA fractions (Table 1) is needed. In contrast, the detection of somatic copy number alterations (SCNA) from low-coverage WGS can be easily performed with low depth and therefore reduced cost. Despite the advantage of low costs and speed, this sequencing strategy limits the detection of SCNAs to those occurring at a minimum of ~5–10% of total ctDNA depending on the amplitude of copy number changes. While high-level gains might be detected down to 1%, heterozygous losses or 1-copy gains are more difficult to catch. Taken together, by being economical and by having the ability to capture the majority of known driver mutations, targeted sequencing still has an advantage over both WES and WGS and might be the preferred method for the detection of low levels of ctDNA, e.g., in early-stage cancer, or monitoring of known clinically relevant mutations. In late stage tumors, tracking the evolution of different cell subclones and the identification of novel—often resistance-conferring or clinically actionable alterations—might be necessary to capture the full extent of tumor heterogeneity. In this context, the greatest benefit of untargeted assessment of tumor-derived changes by WES and WGS is that it does not require individual assays and can be applied for broader clinical utility, independent of recurrent mutations or knowledge about the primary tumor. At the same time, the great potential of ctDNA in providing a more complete picture of the genetic landscape of a tumor than single tissue biopsies aggravates the detection of genetic changes from different locations or clones, as genomic heterogeneity reduces the effective coverage for subclonal alterations and thus the probability of detecting such changes (23). Taken together, a wide variety of options for plasma DNA molecular profiling are available including WGS, WES, large (300–600 gene) panels, small (<50 genes) panels, and hotspots (specific mutations in somatic genes) (13). Hence, the selection of a specific type of genomic profiling should be based on both pre-analytical and analytical factors, considering the fact that increased genomic coverage is detrimental to sensitivity (Figure 1).
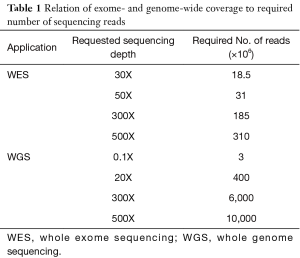
Full table
Whole exome sequencing (WES) of ctDNA
To identify de novo mutations in serial plasma and to track tumor evolution in response to therapy or as a consequence of tumor progression, an untargeted assessment by WES is beneficial over targeted approaches. However, so far there are only a few studies available that have utilized WES for the analysis of ctDNA (selected studies are summarized in Table 2). The first proof-of-concept study for WES of plasma DNA was published in 2013 (9). A total of six patients with advanced breast, ovarian and lung cancers were followed over the course of 1–2 years and WES was performed at multiple time points focusing on samples which were previously analyzed with digital PCR and tagged-amplicon deep sequencing (TAm-Seq) (7) and demonstrated high ctDNA levels (>30%). Starting from less than 5 ng of DNA, the authors report an average of 169 million reads of sequencing per sample, resulting in an average unique coverage depth ranging from 31X to 160X. Due to the high tumor content in the samples, the authors were able to reliably detect genetic changes and achieved a good concordance with respective tumor samples. In addition, a variety of de novo mutations or mutations that were positively selected following treatment and associated with drug resistance were identified. The authors suggested WES as a comprehensive evaluation of clonal genomic evolution associated with treatment response and resistance applicable to patients with high systemic tumour burden. In the following year, a group in Sweden further investigated the utility of using exome sequencing to monitor circulating tumor DNA levels (24). By testing two different library preparation methods for low-input amounts of DNA, the authors assessed the proportion of starting molecules measurable after sequence capture. Despite a significant improvement through an efficient identification of PCR duplicates which enabled massive reduction of background noise, the sensitivity was mainly limited by the poor efficiency of sequence capture (<5%) and the low input amount of DNA (24). Butler et al. conducted WES of cfDNA from two metastatic cancer patients with a higher sequencing depth of 524X and 309X, respectively, and a threshold for variant calling of 1.5% (22). In a sarcoma patient, they were able to reliably detect tumor-specific mutations with an average VAF of 3.7%. Despite an exceptionally high input of 100 ng cfDNA, the enrichment efficiency of 1% was similarly weak as in the above mentioned study. A more recent paper evaluated WES using primary tumors of six NSCLC patients and corresponding cfDNA from 200 µL of serum (25). Libraries were prepared from 10 ng and due to the presence of oligo-nucleosomal laddering, which has previously been associated with the number of CTCs as well as elevated plasma DNA concentrations (10,11), the cfDNA was sonicated. Analysis was performed with a median of 120 million reads leading to a median exome sequencing depth of 68.5X. Library insert sizes of 166 bp indicated that sonication did not further fragment mononucleosomal-derived DNA fragments. The authors did not set any threshold for VAF, except a sequencing depth >10X. Variant calling of matched serum and tissue revealed only a weak concordance with a median of 17.2% (5.2–56.7%), most likely due to the low VAF present in NSCLC in combination with the low sequencing depth (25).
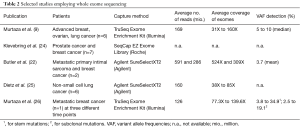
Full table
In another study from Murtaza et al., the authors performed exome sequencing of multiregional tumour biopsies and serial plasma samples and were able to detect and characterise multifocal clonal evolution in a patient with MBC. Not surprisingly, stem mutations (common to all tumour biopsies) had the highest circulating levels in plasma followed by metastatic-clade and private mutations of individual lesions, which reflect the varying responses of different lesions (26). Additional studies to elucidate spatial and temporal heterogeneity and to identify the spectrum and frequency of driver mutations in cancer are ongoing (27,28).
In summary, despite the huge potential for capturing the full extent of genetic evolution at the base pair level, WES of cfDNA comes with inherent limitations and a broad clinical applicability is restricted for several reasons. On the one hand, efficiency of sequence capture is insufficient; on the other hand, the noise levels of sequencing technologies are still too high to achieve suitable sensitivities for the detection of low frequency alleles. Although the introduction of random molecular barcodes can reduce PCR errors, it requires much higher overall numbers of reads to obtain sufficient coverage with consensus reads, which is currently not yet feasible for covering the whole exome. Furthermore, addition of random sequencing to the adapters is likely to complicate adapter blocking during capture, with the risk of decreasing the already low efficiency of capture. Finally, the low fraction of ctDNA and often low overall amounts of total cfDNA requires sampling of larger volumes (>10 mL) of plasma. Therefore, WES approaches are currently only used in research rather than routine clinical settings, at least until sensitivity issues are overcome.
Genome-wide profiling of ctDNA
Genomic instability is one of the hallmarks of cancer (29). Approximately 90% of solid tumors and 50% of blood-related cancers are aneuploid and harbour SCNAs (30). SCNAs include loss of chromosomal material (i.e., deletions), gain of chromosomal material (e.g., duplications), and high-level amplifications with sometimes up to several hundred copies of a relatively small genomic region (3). In contrast to gross deletions and gains which occur mainly due to genetic instability, focal amplifications often occur as a consequence of progression and the selective pressure of therapies. Thus, oncogene amplification occurs late in tumor progression and correlates well with clinical aggressiveness of tumors (31). At these late disease stages, tumors evolve rapidly and, in most cases, it is not possible to retrieve tissue samples to identify novel occurring changes. However, the evolution and the plasticity of tumors can be effectively tracked in plasma using such genome-wide methods (32). Although SCNAs can be established from WES data, compared to WGS, WES introduces more biases and noise that make SCNA detection very challenging. Therefore, in most cases, genome-wide SCNAs are identified from WGS data (Table 3), although a variety of methods including array-CGH and SNP arrays can be used (11,39). As for NGS, read depth (RD)-based approaches are particularly able to detect SCNA regions with an unprecedented resolution. The underlying hypothesis of RD-based methods is that the depth of coverage in a genomic region is correlated with the copy number of the region, e.g., a gain of copy number should have a higher read count than expected (40). For copy number calling, the genome is virtually divided into windows (bins) and reads that are mapped to these genomic regions are counted. After normalization and correction of potential biases (mainly caused by GC content and repeat genomic regions), copy numbers are estimated for each bin. Finally, bins with a similar copy numbers are merged using segmentation algorithms to detect discordant copy number regions (Figure 2) (41).
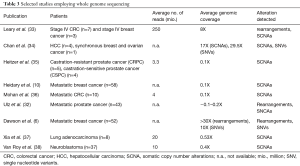
Full table
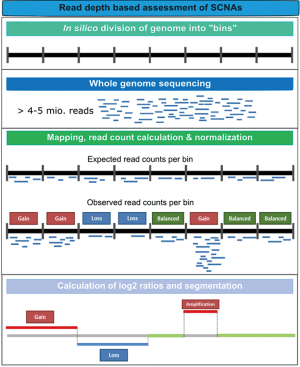
Leary et al. published one of the first studies employing WGS from plasma DNA. The authors aimed to assess aneuploidy and to identify patient-specific rearrangements to design personalized assays for monitoring. To this end, plasma DNA samples were sequenced with an average of 250 million reads. By applying z-score statistics, the authors constructed a log-scale plasma aneuploidy score (PA score) based on the five chromosomes whose arms showed the highest deviations in read counts compared to a healthy control set to distinguish individuals with colorectal and breast cancer from healthy individuals. To identify tumor-derived rearrangement, bioinformatic filters were used that enriched for high-confidence somatic structural alterations while removing germline variants and artefacts. Validated rearrangements were used to develop PCR-based breakpoint-specific assays to accurately determine ctDNA levels. Simulations showed that the sensitivity of detecting tumor-specific SCNAs and rearrangements from WGS data increased proportionally as one over the square root of the number of reads used for analysis, highlighting the fact that sensitivity is largely dependent on the amount of sequence data obtained.
Chan et al. reported the use of WGS to obtain a non-invasive, genome-wide view of cancer-associated copy number variations and mutations in DNA from plasma. While Leary et al. restricted their analysis to chromosome arms, here the authors increased the chromosomal resolution to 1 Mb and a z-score was used to represent increased or decreased 1-Mb windows when compared to a reference group. With respect to sensitivity, the authors came to the same conclusion as Leary et al. that an exponential increase in the number of reads is required to detect copy number changes below 1% tumor DNA. In addition to SCNAs, tumor-associated single nucleotide variants (SNV) were called from WGS data using a sophisticated mathematical algorithm, which takes the actual coverage of the particular nucleotide in the corresponding tumor sequencing data, the sequencing error rate, the maximum false positive rate allowed, and the desired sensitivity for mutation detection into account. The fractional concentrations of tumor-derived DNA were determined based on “genome-wide aggregated allelic loss” (GAAL) analysis, which combined allelic counts for SNVs with identified SCNAs, which is likely to yield a better representation of the actual tumor content in a given sample as individual mutations. By sequencing pre- and post-surgery plasma samples from patients with hepatocellular carcinoma (HCC), breast cancer, and ovarian cancer, the authors showed that their approach is able to qualitatively and quantitatively assess tumor-specific changes.
At the same time, our group developed a very fast and cost-effective approach called plasma-Seq, which is based on low-coverage WGS (0.1–0.2X) and can be performed on the benchtop sequencing MiSeq platform (Illumina, San Diego, CA, USA) (35). Similar to the PA-score, we calculated a genome-wide z-score, which corresponds to the sum of all z-scores from equally-sized 1 Mb-windows and can be used as a general measure of aneuploidy in the sample. Due to the shallow coverage, our approach has a limited analytical sensitivity compared to approaches which use a high depth. Nevertheless, we were able to detect tumor-specific SCNAs with a sensitivity of >80% and specificity of >80%, if >5–10% circulating tumor DNA was present in the samples. Since the detection of SCNA is not only dependent on the sequencing depth but also on the regional copy number, focal amplifications can be detected down to 1% tumor DNA (32). Using plasma-Seq, we were able to monitor genetic evolution including the acquirement of novel copy number changes, such as focal amplifications and chromosomal polysomies in colorectal cancer patients as a response to anti-EGFR therapy (36).
In a recent study, we further developed our algorithm and included calling of focal amplifications, which are thought to most likely contain driver genes. In serial plasma analyses, we observed changes in focal amplifications in 40% of cases, with a mean time interval of 26.4 weeks indicating that late-stage cancers are constantly evolving (32). As these dynamic changes can lead to therapy failure and in parallel to the occurrence of potentially actionable targets, this study together with others highlights the need of comprehensive analyses to effectively track the evolution and the plasticity of tumors. Although WGS sequencing for the detection of SCNA has been applied to a variety of cancers such as breast (6,10), CRC (36), prostate (32,35), lung (37), HCC (34), or neuroblastoma (38), comprehensive studies are still a minority compared to studies analyzing mutation panels or hotspot mutations.
The main reason for this is the lack of sensitivity, as sequencing at affordable costs does not yield informative results below a certain threshold of 5–10% of tumor DNA (2). High levels of ctDNA are mostly seen in metastatic patients, but even these patients sometimes present low levels of ctDNA and approximately 20–30% of samples do not have sufficient amounts of tumor DNA for SCNA analysis (10-12,33). In order to stratify plasma DNA samples based on their ctDNA content, we developed a RD-based approach called mFAST-SeqS using selectively amplified LINE1 sequences (42). While chromosome-arm specific z-scores inform about the copy numbers status on a chromosome arm level, genome-wide z-scores correlate to mutant allele frequencies of somatic mutations and therefore, reflect the tumor fractions (42,43). A distinction of samples with high and low tumor DNA levels can already be achieved with a minimum of 100,000 reads, making this approach a very cheap and fast screening method. The greatest advantage over targeted mutation analysis is certainly the fact that no prior knowledge of tumor-specific alterations is required. Moreover, SCNAs affect a greater fraction of the genome than SNVs; therefore, this method might better reflect the tumor fractions than a single mutation. Finally, since SCNAs are important components of genetic alterations in almost all tumors, this approach can be applied to almost all tumor entities.
Epigenetic profiling of ctDNA
Similar to SCNAs, aberrant DNA methylation and other epigenetic changes are common features of most types of cancers and are thought to occur early in tumor formation (44). Therefore, epigenetic changes have a great potential for early diagnosis of cancer. As with genetic changes, DNA methylation patterns detected in cfDNA are in high concordance with patterns in corresponding primary tumor tissues (45). Also, most studies focusing on epigenetic changes were based on candidate gene approaches (46-48) rather than employing genome-wide profiling (48) (Table 4). The main reasons for this scarcity are technical challenges. Despite the availability of a plethora of methods for genome-wide DNA methylation profiling, such as methylation-specific restriction enzymes, affinity enrichment or bisulfite conversion in combination with microarray or sequencing, the minute amounts of starting material, losses during sample processing (including DNA extraction, bisulfite conversion and library preparation) as well as poor recovery rates for methylation enrichment hinder broad application. Warton et al. addressed these challenges and recently reported a detailed protocol for plasma DNA extraction and enrichment of methylated sequences followed by NGS (49).
Full table
In 2013, the group of Dennis Lo further developed their WGS-based approach and explored the utility of bisulfite sequencing for the detection genome-wide hypomethylation and SCNAs as a marker for cancer (50). First, plasma DNA samples obtained from 26 hepatocellular carcinoma (HCC) patients and 32 healthy subjects were used to assess the performance of plasma hypomethylation and SCNA detection based on reads count analysis (50). By combining both types of alterations, a sensitivity and specificity of 69% and 94%, respectively, could be achieved for the detection of HCC. Second, 20 other cancer samples including breast cancer, lung cancer, nasopharyngeal cancer, smooth muscle sarcoma, and neuroendocrine cancer were analyzed. As expected, patients with metastatic stages had the highest percentages of bins showing plasma hypomethylation and SCNAs reflecting higher tumor DNA levels. While subsampling of reads from an average of 93 million to 10 million dramatically affected the diagnostic performance of SCNAs (drop from 71% to 39%), it had only a minor influence on the hypomethylation analysis. In addition to a diagnostic application, the author showed that this combined approach had utility for monitoring hepatocellular carcinoma patients following tumor resection and for detecting residual disease (50).
Using MethylCap-seq, Zhao et al. analyzed genome-wide cfDNA methylation profiles of healthy controls, patients with chronic hepatitis B infection or liver cirrhosis, and HCC (51) and identified potential methylation markers for the early detection of HCC. Data mining revealed the presence of 240, 272 and 286 differentially methylated genes (DMGs) corresponding to the early, middle and late stages of HCC progression, respectively indicating that the dynamic features of cfDNA methylation coincided with the natural course of HBV-related HCC development (51).
Another study investigated the use of circulating DNA methylation changes for prediction of MBC (52). Using whole genome bisulfite sequencing (average sequencing depth of 11X), ~5×106 differentially methylated CpG loci (DML) in MBC compared with healthy individuals or disease-free survivors were identified. Consistent with common global hypomethylation in cancer, most differentially methylated loci (90%) were hypomethylated and only a small set of DML could be attributed to focal CpG island hypermethylation. To identify potential biomarkers of MBC, data mining was performed and 21 novel hotspots were identified within CpG islands which differed most from healthy individuals or disease-free survivors, which might be used for stratification of patients who are at a high risk of recurrence and who could benefit from additional therapy.
Zhai et al. compared the concordance of genome-wide methylation patterns between tumor tissues and corresponding sera to assess the performance of genome-wide methylation profiles in differentiating esophageal adenocarcinoma, their precursor the Barrett esophagus, and controls. Using the Infinium HumanMethylation27 BeadChip, the authors observed high concordance (r=92) of aberrantly methylated loci of serum DNA and the respective tumors. Clustering analyses showed that 911 loci perfectly discriminated between EA and control samples, 544 loci separated esophageal adenocarcinoma from Barrett esophagus samples, and 46 loci distinguished Barrett esophagus from control samples, suggesting that DML established from plasma DNA may be valuable biomarkers for early detection of esophageal adenocarcinoma.
Recently, a novel DNA methylation analysis technique called MCTA-Seq (methylated CpG tandems amplification and sequencing) for genome-wide detection of hypermethylated CGIs in plasma DNA was reported (54). This procedure is based on a single-tube three-step amplification of very short DNA fragments adjacent to the methylated CGCGCGG sequences from bisulfite-treated DNA. The authors identified 2,166 differentially hypermethylated CGIs in plasma of HCC patients, of which only a very small portion of CGIs hypermethylated in the HCC tissues were good markers for detecting HCC in blood. Among these markers, 4 (RGS10, ST8SIA6, RUNX2 and VIM) were mostly specific for HCC detection, while the other 15 were already hypermethylated in the normal liver and might be used to assess tissue contributions in plasma DNA (54).
As DNA methylation presents both tissue- and tumor-specific patterns, it is a particularly attractive marker with broad application in diagnostics. In addition to genome-wide and locus-specific alterations, it possible to determine the tissue-of origin of circulating fragments (55), which may indicate that a cancer is located in or originates from a specific tissue.
Moving beyond SCNA and SNV
Recently, comprehensive datasets were used to shed light onto the yet unknown biology of plasma-derived DNA and to further assess its use as a biomarker. Apart from SNVs and SCNAs, the size of DNA fragments has been suggested to be an important parameter to distinguish tumor-derived DNA from normal DNA. Using WGS for size profiling, Jiang et al. demonstrated slightly shorter size fragments, which preferentially carried the SCNAs in HCC patients compared to controls (56). A study of plasma DNA in xenografted rats also revealed shorter fragment lengths of tumor-derived fragments than the background rat cell-free DNA. A similar shift in the fragment length of ctDNA in humans with melanoma and lung cancer was identified when compared to healthy controls (57). A selection of DNA fragments between 90-150 bp before analysis yielded enrichment of mutated a DNA fraction of up to 11-fold (58). Moreover, efforts are being made to develop library preparation protocols that enrich for shorter fragments (59). However, using this protocol for genome-wide analysis of plasma DNA samples from metastatic cancer patients, we did not observe an enrichment of tumor-derived fragments, despite a significant enrichment of short DNA fragments (60). Moreover, we observed a correlation of the presence of larger fragment sizes in the range of di- and tri-nucleosomal fragments and high levels of tumor DNA (10,11). These data still indicate a controversy on whether the higher or lower integrity of cfDNA is associated with cancer and suggest that there might be different mechanisms and dynamics for DNA degradation.
In addition to the nature of circulating DNA fragments, their origin is increasingly becoming the focus of research and it has been demonstrated that cfDNA originates from different tissues of the body (61-65). Both methylation and nucleosome occupancy patterns were able to deconvolute tissue of origin, although results were not fully consistent within the different studies (64,65). However, there is a common understanding that cfDNA is primarily derived from apoptosis of normal cells of the hematopoietic lineage and other solid tissues contribute only to a small part of cfDNA. As available studies lack either a prediction method or systematic performance evaluations, Kang et al. developed a probabilistic method using genome-wide DNA methylation data to predict presence and location of a tumor (62). Nevertheless, the potential of tissue-specific cfDNA signals need to be further investigated.
Another potentially useful parameter which can be extracted from plasma DNA is that of nucleosome occupancy patterns. The Shendure group investigated nucleosome phasing patterns from WGS data and were able to track the tissue(s)-of-origin of plasma DNA (64). Ivanov et al. took a step forward and tested for the association of these genomic coordinates with the relative strength and the patterns of gene expression in cfDNA samples (66). Since the nucleosome density varies at transcription start sites (TSS) of actively transcribed and non-expression genes, we investigated whether different abundances based on depth analyses of DNA fragments at TSS can inform about the expression status of genes (67). We showed that the plasma DNA read depth patterns from healthy donors reflected the expression signature of hematopoietic cells and, moreover, we were able to classify expressed cancer driver genes in regions with somatic copy number gains with high accuracy in metastatic cancer patients with high tumor load (67).
Overall these data indicate that besides common genetic and epigenetic alterations additional information is hidden in plasma DNA and comprehensive analyses can provide functional and biological knowledge.
Conclusions
Although the advent of NGS technology has accelerated the path to precision medicine by being able to characterize many tumor types at the molecular level, inherent limitations to detection and multiplexing approaches have forced clinicians and researchers alike to adopt either targeted or untargeted approaches based on the main clinical question at hand. WGS represents perhaps the most straightforward application of NGS in general, as no enrichment step is required. This untargeted method does not require pre-knowledge of the tumor and can provide high-resolution information regarding SCNAs and chromosomal rearrangements. Tumor heterogeneity is best captured at the whole-genome level, as a global approach will not targetedly exclude potential regions of interest harbouring genetic alterations and allows for unbiased evaluation of the entire tumor content. This is of particular importance for detecting and tracking developing subclones and identifying potentially clinically druggable mutations as recently it was demonstrated that 86% of tumors across 12 cancer types had at least two clones (68).
Furthermore, this method can not only easily be conducted at low depth, but it is a relatively quick approach, thus reducing overall costs associated with sequencing. However, despite these financial advantages and unbiased testing, this method is unfortunately currently limited to samples with a presence of 5–10% of total ctDNA, dependent on copy number amplitudes. Similar restrictions apply to WES. These limitations speak to the advantage of targeted sequencing approaches over WGS/WES, which have the potential of capturing major known driver mutations, an application which is of course high priority for treating physicians in the clinic. Moreover, targeted sequencing has greater potential for the detection of lower levels of ctDNA which could be present in patients with early-stage disease, thus emphasizing utility in pre-screening efforts and the possibility of early detection of resistance or relapse. A multitude of factors influence the decision for selecting genomic profiling approaches and the best clinical strategy depends on the tumor information or lack thereof in a given situation. As sequencing technology and the understanding of ctDNA continues to evolve in parallel, the variety of options for molecular profiling of plasma DNA will also continue to increase and adapt to patient-specific scenarios, thus bringing the idea of precision medicine ever closer.
Acknowledgments
Funding: The work in our laboratory is supported by CANCER-ID, a project funded by the Innovative Medicines Joint Undertaking (IMI JU); the Austrian National Bank [ÖNB, grant# 16917]; the Austrian Science Fund [FWF, grant# P28949-B28]; and by the BioTechMed-Graz flagship project “EPIAge”.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Heidi Schwarzenbach) for the series “Technologies in Liquid Biopsies - Potential applications in Medicine” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.10.11). The series “Technologies in Liquid Biopsies - Potential applications in Medicine” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cheng F, Su L, Qian C. Circulating tumor DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget 2016;7:48832-41. [PubMed]
- Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem 2015;61:112-23. [Crossref] [PubMed]
- Heitzer E, Ulz P, Geigl JB, et al. Non-invasive detection of genome-wide somatic copy number alterations by liquid biopsies. Mol Oncol 2016;10:494-502. [Crossref] [PubMed]
- Wan JC, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]
- Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531-48. [Crossref] [PubMed]
- Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199-209. [Crossref] [PubMed]
- Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012;4:136ra68 [Crossref] [PubMed]
- Olsson E, Winter C, George A, et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol Med 2015;7:1034-47. [Crossref] [PubMed]
- Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [Crossref] [PubMed]
- Heidary M, Auer M, Ulz P, et al. The dynamic range of circulating tumor DNA in metastatic breast cancer. Breast Cancer Res 2014;16:421. [Crossref] [PubMed]
- Heitzer E, Auer M, Hoffmann EM, et al. Establishment of tumor-specific copy number alterations from plasma DNA of patients with cancer. Int J Cancer 2013;133:346-56. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24 [Crossref] [PubMed]
- Perakis S, Speicher MR. Emerging concepts in liquid biopsies. BMC Med 2017;15:75. [Crossref] [PubMed]
- Perakis S, Auer M, Belic J, et al. Advances in Circulating Tumor DNA Analysis. Adv Clin Chem 2017;80:73-153. [Crossref] [PubMed]
- Rehm HL, Bale SJ, Bayrak-Toydemir P, et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med 2013;15:733-47. [Crossref] [PubMed]
- Drilon A, Wang L, Arcila ME, et al. Broad, Hybrid Capture-Based Next-Generation Sequencing Identifies Actionable Genomic Alterations in Lung Adenocarcinomas Otherwise Negative for Such Alterations by Other Genomic Testing Approaches. Clin Cancer Res 2015;21:3631-9. [Crossref] [PubMed]
- Kinde I, Wu J, Papadopoulos N, et al. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A 2011;108:9530-5. [Crossref] [PubMed]
- Schmitt MW, Kennedy SR, Salk JJ, et al. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A 2012;109:14508-13. [Crossref] [PubMed]
- Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547-55. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014; [Epub ahead of print]. [Crossref] [PubMed]
- Shu Y, Wu X, Tong X, et al. Circulating Tumor DNA Mutation Profiling by Targeted Next Generation Sequencing Provides Guidance for Personalized Treatments in Multiple Cancer Types. Sci Rep 2017;7:583. [Crossref] [PubMed]
- Butler TM, Johnson-Camacho K, Peto M, et al. Exome Sequencing of Cell-Free DNA from Metastatic Cancer Patients Identifies Clinically Actionable Mutations Distinct from Primary Disease. PLoS One 2015;10:e0136407 [Crossref] [PubMed]
- Bashir A, Volik S, Collins C, et al. Evaluation of paired-end sequencing strategies for detection of genome rearrangements in cancer. PLoS Comput Biol 2008;4:e1000051 [Crossref] [PubMed]
- Klevebring D, Neiman M, Sundling S, et al. Evaluation of exome sequencing to estimate tumor burden in plasma. PLoS One 2014;9:e104417 [Crossref] [PubMed]
- Dietz S, Schirmer U, Merce C, et al. Low Input Whole-Exome Sequencing to Determine the Representation of the Tumor Exome in Circulating DNA of Non-Small Cell Lung Cancer Patients. PLoS One 2016;11:e0161012 [Crossref] [PubMed]
- Murtaza M, Dawson SJ, Pogrebniak K, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 2015;6:8760. [Crossref] [PubMed]
- Chicard M, Daage LC, Clement N, et al. Abstract 4952: Whole exome sequencing of circulating tumor DNA highlights spatial and temporal tumor heterogeneity in neuroblastoma. Proceedings: AACR Annual Meeting 2017; April 1-5, 2017; Washington, DC.
- Beltran H, Romanel A, Casiraghi N, et al. Whole exome sequencing (WES) of circulating tumor DNA (ctDNA) in patients with neuroendocrine prostate cancer (NEPC) informs tumor heterogeneity. J Clin Oncol 2017;35:abstr 5011.
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010;463:899-905. [Crossref] [PubMed]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature 1998;396:643-9. [Crossref] [PubMed]
- Ulz P, Belic J, Graf R, et al. Whole-genome plasma sequencing reveals focal amplifications as a driving force in metastatic prostate cancer. Nat Commun 2016;7:12008. [Crossref] [PubMed]
- Leary RJ, Sausen M, Kinde I, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med 2012;4:162ra154 [Crossref] [PubMed]
- Chan KC, Jiang P, Zheng YW, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem 2013;59:211-24. [Crossref] [PubMed]
- Heitzer E, Ulz P, Belic J, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med 2013;5:30. [Crossref] [PubMed]
- Mohan S, Heitzer E, Ulz P, et al. Changes in colorectal carcinoma genomes under anti-EGFR therapy identified by whole-genome plasma DNA sequencing. PLoS Genet 2014;10:e1004271 [Crossref] [PubMed]
- Xia S, Huang CC, Le M, et al. Genomic variations in plasma cell free DNA differentiate early stage lung cancers from normal controls. Lung Cancer 2015;90:78-84. [Crossref] [PubMed]
- Van Roy N, Van der Linden M, Menten B, et al. Shallow whole genome sequencing on circulating cell-free DNA allows reliable non-invasive copy number profiling in neuroblastoma patients. Clin Cancer Res 2017;23:6305-14. [Crossref] [PubMed]
- Shaw JA, Page K, Blighe K, et al. Genomic analysis of circulating cell-free DNA infers breast cancer dormancy. Genome Res 2012;22:220-31. [Crossref] [PubMed]
- Yoon S, Xuan Z, Makarov V, et al. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res 2009;19:1586-92. [Crossref] [PubMed]
- Magi A, Tattini L, Pippucci T, et al. Read count approach for DNA copy number variants detection. Bioinformatics 2012;28:470-8. [Crossref] [PubMed]
- Belic J, Koch M, Ulz P, et al. Rapid Identification of Plasma DNA Samples with Increased ctDNA Levels by a Modified FAST-SeqS Approach. Clin Chem 2015;61:838-49. [Crossref] [PubMed]
- Belic J, Koch M, Ulz P, et al. mFast-SeqS as a Monitoring and Pre-screening Tool for Tumor-Specific Aneuploidy in Plasma DNA. Adv Exp Med Biol 2016;924:147-55. [Crossref] [PubMed]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011;11:726-34. [Crossref] [PubMed]
- Warton K, Samimi G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Front Mol Biosci 2015;2:13. [Crossref] [PubMed]
- Begum S, Brait M, Dasgupta S, et al. An epigenetic marker panel for detection of lung cancer using cell-free serum DNA. Clin Cancer Res 2011;17:4494-503. [Crossref] [PubMed]
- Wong IH, Johnson PJ, Lai PB, et al. Tumor-derived epigenetic changes in the plasma and serum of liver cancer patients. Implications for cancer detection and monitoring. Ann N Y Acad Sci 2000;906:102-5. [Crossref] [PubMed]
- Fackler MJ, Lopez Bujanda Z, Umbricht C, et al. Novel methylated biomarkers and a robust assay to detect circulating tumor DNA in metastatic breast cancer. Cancer Res 2014;74:2160-70. [Crossref] [PubMed]
- Warton K, Lin V, Navin T, et al. Methylation-capture and Next-Generation Sequencing of free circulating DNA from human plasma. BMC Genomics 2014;15:476. [Crossref] [PubMed]
- Chan KC, Jiang P, Chan CW, et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci U S A 2013;110:18761-8. [Crossref] [PubMed]
- Zhao Y, Xue F, Sun J, et al. Genome-wide methylation profiling of the different stages of hepatitis B virus-related hepatocellular carcinoma development in plasma cell-free DNA reveals potential biomarkers for early detection and high-risk monitoring of hepatocellular carcinoma. Clin Epigenetics 2014;6:30. [Crossref] [PubMed]
- Legendre C, Gooden GC, Johnson K, et al. Whole-genome bisulfite sequencing of cell-free DNA identifies signature associated with metastatic breast cancer. Clin Epigenetics 2015;7:100. [Crossref] [PubMed]
- Zhai R, Zhao Y, Su L, et al. Genome-wide DNA methylation profiling of cell-free serum DNA in esophageal adenocarcinoma and Barrett esophagus. Neoplasia 2012;14:29-33. [Crossref] [PubMed]
- Wen L, Li J, Guo H, et al. Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Cell Res 2015;25:1376. [Crossref] [PubMed]
- Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer 2003;3:253-66. [Crossref] [PubMed]
- Jiang P, Chan CW, Chan KC, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A 2015;112:E1317-25. [Crossref] [PubMed]
- Underhill HR, Kitzman JO, Hellwig S, et al. Fragment Length of Circulating Tumor DNA. PLoS Genet 2016;12:e1006162 [Crossref] [PubMed]
- Mouliere F, Piskorz AM, Chandrananda D, et al. Selecting Short DNA Fragments In Plasma Improves Detection Of Circulating Tumour DNA. bioRxiv preprint first posted online May. 5, 2017; doi: http://dx.doi.org/
10.1101/134437 - Burnham P, Kim MS, Agbor-Enoh S, et al. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci Rep 2016;6:27859. [Crossref] [PubMed]
- Moser T, Ulz P, Zhou Q, et al. Single-Stranded DNA Library Preparation Does Not Preferentially Enrich Circulating Tumor DNA. Clin Chem 2017;63:1656-9. [Crossref] [PubMed]
- Thierry AR, El Messaoudi S, Gahan PB, et al. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev 2016;35:347-76. [Crossref] [PubMed]
- Kang S, Li Q, Chen Q, et al. CancerLocator: non-invasive cancer diagnosis and tissue-of-origin prediction using methylation profiles of cell-free DNA. Genome Biol 2017;18:53. [Crossref] [PubMed]
- Lehmann-Werman R, Neiman D, Zemmour H, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci U S A 2016;113:E1826-34. [Crossref] [PubMed]
- Snyder MW, Kircher M, Hill AJ, et al. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell 2016;164:57-68. [Crossref] [PubMed]
- Sun K, Jiang P, Chan KC, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A 2015;112:E5503-12. [Crossref] [PubMed]
- Ivanov M, Baranova A, Butler T, et al. Non-random fragmentation patterns in circulating cell-free DNA reflect epigenetic regulation. BMC Genomics 2015;16:S1. [Crossref] [PubMed]
- Ulz P, Thallinger GG, Auer M, et al. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat Genet 2016;48:1273-8. [Crossref] [PubMed]
- Andor N, Graham TA, Jansen M, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 2016;22:105-13. [Crossref] [PubMed]


