
Clinical relevance of size selection of circulating DNA
Introduction
In 1948, circulating cell-free DNA (cfDNA) was primarily detected and described by Mandel and Metais (1). It is passively released by cell death (2) from all cell types (3) and actively by cell secretion (4). It can be found in all body fluids, such as blood (5), saliva (6), amniotic fluids (7) and urine (8,9). The biggest progress in cfDNA investigation was the moment of the discovery that gender of the fetus can be identified by PCR amplification of the Y-chromosome sequences in cfDNA from maternal blood (10). Now, non-invasive diagnosis of fetal genetic disorders is successively applied, worldwide (11-13). Since cfDNA provides information on tumor genetic and epigenetic changes in nuclear DNA, it can also serve as a powerful tool for monitoring patients with different tumor types (14). For example, aberrant DNA methylation, mutations, copy number aberrations (CNA) and chromosomal rearrangements, resulting in inactivation of tumor suppressor genes and activation of oncogenes, can be found in cfDNA isolated from cancer patients and can be utilized for detection, monitoring and prognosis of cancer patients (15,16).
An important aspect in the research field of cfDNA is the determination of their length, to identify its origin. It is known that cfDNA fragment lengths in various pathological/physiological stages differ from those of normal cfDNA fragments (12,15-19). Therefore, analyses of cfDNA length fragment distribution may contribute to advance diagnostic tools. Disease-associated cfDNA is usually contaminated with normal wild-type DNA. In this regard, it is important for an efficient analysis to separate disease-associated cfDNA from background cfDNA (i.e., fetal from maternal or tumor from non-tumor cfDNA). To date, an exact fractionation in disease-associated and normal cfDNA is not possible, since cfDNA size distribution is not sharp defined and may vary in different diseases. Moreover, technological limitations may also play a role.
Characterization of cfDNA
In the blood circulation, cfDNA exists mostly as nuclear, histone complexed DNA (20). This complex, called nucleosome, protects cfDNA from enzymatic degradation. Nucleosomal DNA consists of ~146 bp molecule bound electrostatically to the histone core and the 20-bp linker connecting the nucleosomes (21). The most common size of histone complexed DNA is ~166 bp, and while analyzing cfDNA size distribution, a specific ladder pattern of multiplies of this size can be detected in the human blood (15,22-24). This periodic pattern is found in plasma DNA from all kind of subjects, such as pregnant women (17), organ transplant recipient (18), and cancer patients (15) (Table 1). It is caused by the apoptotic inter-nucleosomal DNA cleavage mediated by the caspase-activated DNase in dying cells and the DNase II during phagocytosis (15,21). Fragments shorter than ~166 bp can be the result of the linker trimming (17) or the degradation of the non-nucleosomal cfDNA. The general size of circulating nuclear DNA in bloodstream of healthy individuals have been described to vary mainly between 70 and 200 bp (14,18). In contrast, long cfDNA molecules in size of more than 10,000 bp are also found in the blood and stem from necrotic cells (20,22). Thus, the variety of the different cfDNA length depends on the frequency of cell apoptosis and necrosis, as well as different metabolic processes out of the cells.
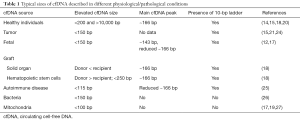
Full table
Of note is that cfDNA may play a role in the immune system, and in certain circumstances, cfDNA is immunogenic. In particular, shorter, non-histone complexed cfDNA are assumed to activate the immune system response (28,29). So, the induction of inflammatory reactions mediated by cfDNA may occur by its binding to the toll-like receptor 9 (TLR9). For example, fetal cfDNA may interact with TLR9 and result in the activation of mothers immune reactivation (30). Moreover, DNA can be actively released by blood lymphocytes (31). In vitro studies showed that this cfDNA can enter other cells and be expressed there (32). Also antibodies induced by cfDNA can enter cells, leading to cell-cycle arrest or apoptosis (REF) (33). These findings suggest that assessment of cfDNA levels and its characteristics may be useful for the identification of different inflammatory processes, and provide evidence that cfDNA is a potential activator of the immune system in response to disorders.
cfDNA size profiles in different pathological/physiological conditions
Tumor
In the blood of cancer patients, both, tumor and non-tumor derived cfDNA can be found. The amount of circulating tumor DNA (ctDNA) in the whole pool of cfDNA may vary significantly among the patients, from 0.01% to 90%, and be related to tumor size (23).
It is assumed that necrosis accompanies cancer development, whereas phagocytosis and apoptosis are defense responses of the organism and may result in the destruction of cancer cells as well as the surrounding non-cancer tissues (14,22). It is still being discussed whether ctDNA mainly displays lower or higher fragmentation. Several studies demonstrated that the mean length of cfDNA increases in patients with non-malignant tumor (15,16,20), while other laboratories showed a higher fragmentation of ctDNA comparing it to cfDNA originating from healthy cells (22,34). This finding was confirmed by another group that investigated ctDNA fragment lengths in patients with hepatocellular carcinoma (HCC). HCC patients displayed increased fractional concentrations of more fragmented ctDNA in the plasma. The researchers also found that the number of DNA fragments shorter than 150 bp increased directly proportional to ctDNA concentration. Thus, smaller proportions of fragments longer than 180 bp were found with increased ctDNA concentrations. Moreover, plasma DNA carrying CNA were shorter than cfDNA without these deviations (15). In a more recent study, ctDNA from patients with different solid tumors was demonstrated to be always gently shorter than DNA derived from healthy cells. Moreover, the separation of shorter cfDNA fragments visibly increased with the proportion of ctDNA. Tumor specific mutant allele occurred more frequently in the fraction of 132–145 bp cfDNA than in the fraction of 165 bp DNA (34).
These findings show that cfDNA size selection might be useful as an additional diagnostic tool for detection of cancer-derived changes in patient’s plasma.
Pregnancy
Overall, the size of cfDNA increases during pregnancy, whereas fetal cfDNA is significantly shorter than maternal cfDNA (35,36). Generally, the length of fetal cfDNA in maternal plasma is shorter than 500 bp and the major portion is shorter than 300 bp (25,26). It was also reported that fetal cfDNA encompasses a higher proportion of cfDNA fragments smaller than 150 bp in comparison with maternal DNA (12,17,18). There is a notable amount of maternal high-molecular weight cfDNA fragments, which are longer than 10,000 bp (35). Size analysis of cfDNA from maternal plasma revealed that the characteristic peak of maternal cfDNA at ~166 bp is decreased, and there is an additional peak at 143 bp. This peak is probably caused by trimming of a ~20 bp linker segment of a 166 bp fragment, leaving a remnant of a mono-chromatosome of ~146 bp length which is wrapped around the histone core. Size distributions smaller than 143 bp of fetal and maternal DNA showed a length pattern with systematical decreases by 10 bp (17).
These size-based analyses of maternal plasma DNA and the discovery that fetal cfDNA is shorter than maternal cfDNA allow detecting chromosomal aneuploidies, including monosomy X and trisomies (12,13). Moreover, studies on size distributions of cfDNA in pregnant women can be successfully used to detect fetal sub-chromosomal CNA. Finally, the combination of count- and size-based methods can help to understand if CNA occurs in fetus, mother or in both species (13).
Since in maternal plasma, a median of 10–20% of cfDNA is fetus-originated (27,36-38), a size-based separation of fetal cfDNA from background maternal cfDNA might contribute to improve the noninvasive prenatal diagnostics (12,39).
Transplantation
In recipients of bone marrow or solid organ transplantation, different length patterns of cfDNA originating from transplanted organs have been observed. Based on analysis of plasma cfDNA of hematopoietic stem cell transplants recipients, it was showed that the size of recipient’s cfDNA is usually significantly shorter than donor-derived cfDNA, resulting in a median difference of almost 15%. Thus, it appears that non-hematopoietically-derived DNA is shorter than DNA of hematopoietical origin. In details, the entire pool of (recipient- and donor-derived) cfDNA contained relatively short cfDNA fragments, since more than 98% of cfDNA was shorter than 250 bp. There were a characteristic peak at 166 bp length and 10 bp periodical peaks in sizes smaller than 143 bp. The 166-bp peak of recipient’s cfDNA was reduced. This can be explained by the fact that the majority (around 85%) of plasma cfDNA was donor-derived and its overall size is somewhat longer.
In contrast to bone marrow transplants, plasma of a recipient of liver transplant contained mostly cfDNA of recipient’s origin (over 70%), which was found to be longer than donor’s cfDNA. Since the liver is a non-hematopoietic organ in adults, the liver transplant releases only non-hematopoietic DNA, confirming the observation that cfDNA of hematopoietic origin is somewhat longer than of non-hematopoietic origin (18).
Autoimmune diseases
Already in 1990, it was reported that cfDNA isolated from systemic lupus erythematosus (SLE) patients is not randomly fragmented, but represents a specific ladder-like pattern of multiple 200 bp lengths, suggesting that cfDNA is bound at histones and digested by DNases during apoptosis (40). Twenty-four years later, plasma cfDNA of SLE patients was described to have a reduced, characteristic 166-bp peak and have a higher proportion of cfDNA fragments smaller than 115 bp. The median proportion of shorter fragments (<115 bp) was 10% in the group of healthy individuals, and increased to 14% in the inactive SLE group and to 31% in the active SLE group. There was also a positive relationship between the proportion of cfDNA fragments shorter than 115 bp, SLE activity and the anti-dsDNA antibody level, supporting the fact that antibodies bind preferentially to shorter cfDNA fragments. Fragmentation, antibody affinity and SLE activity increased in line with the cfDNA hypomethylation, since hypomethylated cfDNA is bound less tightly with nucleosomes, and therefore is more susceptible for enzymatic activity and antibody binding (29).
Infections
Size distribution of microbial and mitochondrial circulating cell-free DNA (mcfDNA) is expected to be similar due to their common evolution history. Recently, plasma samples from sepsis patients were analyzed by massively parallel sequencing (MPS) that detected microbial DNA of Propionibacterium acnes, Streptococcus agalactiae and Epstein-Barr virus (EBV). Interestingly, cfDNA of both bacteria were more fragmented than human cfDNA, with a majority of cfDNA fragments shorter than 150 bp, whereas EBV-derived DNA had a characteristic peak in a similar size to human cfDNA (41). A further study of cfDNA from lung transplant recipients demonstrated that bacterial DNA detected in the blood circulation of those patients is also more fragmented when comparing to human nuclear cfDNA. Therefore, the analysis of shorter microbial cfDNA and consequently, size-based cfDNA separation might be of interest in transplant recipients, who are undergoing intensive immunosuppressive therapies, to decrease the risk of transplant rejection and opportunistic infections (42).
Circulating mitochondrial DNA
The pattern of mcfDNA fragments is different from that of nuclear cfDNA. In mcfDNA, the peak of 166 bp characteristic for nuclear DNA is not observed, and the overall size distribution of mcfDNA is smaller than of nuclear genomic cfDNA (15,19,42). Its highest concentration was primarily reported in a size of 130–140 bp, as measured by standard MPS (15). However, newest investigations with optimized protocols of MPS showed a highest abundance of mcfDNA fragments smaller than 100 bp (19,42), but a lack of a nucleosomal mcfDNA distribution pattern in a periodical size of 10-bp, typical for histone-bound cfDNA. The smaller overall length distribution of mcfDNA is a result of enzymatic degradation, which occurs much easier due to the lack of histone protection of mcfDNA (15,17). Moreover, in a previous study on HCC patients, the concentrations of mcfDNA in plasma of these patients were higher than those of healthy patients (0.0014% and 0.00045%, respectively, as measured by standard MPS) (15). However, this firstly described overall abundance of mcfDNA seemed to be underestimated. In fact, using an improved quantification method of shorter mcfDNA fragments by digital PCR (dPCR), and due to the presence of 50–4,000 of mitochondrial genomes per cell, an abundance of mcfDNA 56-fold higher than that of nuclear genomic DNA was discovered. In addition, the same study showed that mcfDNA released from graft is slightly shorter than mcfDNA originating from a recipient (42). Finally, another study showed that in serum of patients with testicular germ cell cancer, the levels of 79- and 220-bp mcfDNA fragments were elevated in comparison with healthy controls, and the proportion of 79-bp length mcfDNA molecules was higher than that of 220-bp mcfDNA molecules, indicating the lower integrity of mcfDNA in this pathological condition (43).
cfDNA size determination and separation methods, and their utility
The selection of a defined size range of cfDNA is an important preliminary step prior to massive parallel sequencing (19,44-46). Furthermore, size-dependent separation of cfDNA fragments have been already implemented in the development and improvement of non-invasive prenatal detection of fetal genetic disorders (35,36). Therefore, it is expected that the enrichment of cfDNA according to their size may also contribute to improve diagnostic tests, e.g., the detection of genetic aberrations in blood of tumor patients. In this regard, several techniques have been developed and are summarized below (Figure 1).
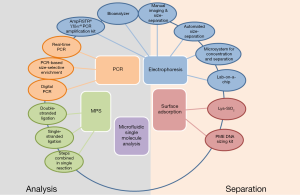
Electrophoresis-based techniques
Gel electrophoresis imaging and size-separation
The easiest technique to determine the length of cfDNA is the conventional gel electrophoretic separation (34,35). However, the main disadvantage of this method is its low sensitivity. For example, more than a decade ago size-fractionated DNA was still isolated from a gel to enrich fetal cfDNA from maternal plasma (35,47). At that time, Li et al. found that the highest proportion of fetal cfDNA was smaller than 300 bp. Moreover, the gel electrophoresis-based enrichment of fetal cfDNA also facilitated the analysis of maternally and paternally inherited DNA polymorphisms (35).
An advanced alternative to the standard gel electrophoresis is an automated gel separation and size selection. Here, an instrument is loaded with disposable cassette containing prefabricated agarose gel. The electrophoresis of cfDNA samples is running until a laser detector determines the time point of collecting the assembled cfDNA referring to the previously established collection size range. This sizing method can be dedicated for the selection of DNA libraries for new generation sequencing (NGS), and was recently, successfully used to separate cfDNA in size of 90–150 bp and cfDNA libraries in size of 150–190 bp, to enrich ctDNA and short mcfDNA fragments, respectively, from the total cfDNA pool prior to sequencing (19,46).
Bioanalyzer
The Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) is a platform performing a microchip-based capillary electrophoresis of nucleic acids and proteins. Its software automatically analyzes and adjusts the cfDNA length of the defined regions, and calculates the average size (bp), size distribution in coefficient of variance (CV, %), concentration (pg/µL), percentage of total DNA, and molarity (pmol/L) for each designated cfDNA region. Using this method, size distribution and concentration of cfDNA can be measured in a digital format. The relatively short time for the performance of an experiment and the input of low volumes of purified cfDNA samples make this technique advantageous (2,12,34,48-50).
In this regard, plasma cfDNA of colorectal cancer (CRC) patients was tested with this platform. Twenty-one of 32 CRC patients had a similar size distribution of cfDNA to the control group of healthy volunteers with a pronounced peak in a size of 85–230 bp. In the remaining 11 CRC patients an additional peak in a size of 240–400 bp was observed. These patients had also approximately 6-times higher concentrations of cfDNA than the other CRC patients. These high cfDNA concentrations positively correlated with the occurrence of circulating tumor cell and in some cases with an higher percentage of KRAS mutated cfDNA (49). A year later, the same researchers also determined the cfDNA size profiles of women with metastasized breast cancer. They observed an additional cfDNA peak of longer cfDNA fragments in 25 of 57 tested patients, and reported that while the peak of shorter cfDNA fragments likely corresponds to apoptotic mono-nucleosomal DNA, the second peak might represent 2- or 3-nucleosomal cfDNA fragments released by phagocytosis (50).
Moreover, the size distribution of adaptor-ligated cfDNA library enriched by PCR can be analyzed prior to sequencing by the Bioanalyzer (12,34). Yu et al. analyzed cfDNA libraries of pregnant women. To get accurate cfDNA lengths, the length of the adaptors was subtracted from all cfDNA fragments. Then, the ratio of the areas of 78–143 and 163–168 bp was calculated. The fetal cfDNA fraction in maternal plasma could be estimated based on the fact that cfDNA contained increased amounts of molecules smaller than 150 bp and a decreased peak at 166 bp (12).
Finally, size analysis by the Bioanalyzer can also be utilized to check the purity of cfDNA for the absence of cell DNA, which is a high-molecular weight DNA (44).
Microsystem for concentration and size separation of fetal cfDNA
In 2009, Hahn et al. introduced a micro-device that encompasses electrokinetic trapping, transient isotachophoresis and capillary electrophoresis for automated enrichment and size-depending separation of fetal cfDNA. In the first step, cfDNA isolated from maternal plasma is applied in the polymeric microsystem, in which by means of electrokinetic trapping, cfDNA is captured and subsequently pre-concentrated. In the second step, by applying transient isotachophoresis cfDNA is put in the matrix of polymerized acrylamide in the separation channel. After exchanging buffers in the system, cfDNA is size-separated by performing capillary electrophoresis, and each 30 seconds 2-µL fractions are manually collected from recovery outlet. The device is disposable and easily operated. The microsystem was able to separate DNA fragments of the 100-bp DNA ladder into four fractions that contained cfDNA fragments of 100–300, 300–500, 500–600 and >600 bp collected between 9.0–10.0, 10.5–11.5, 12.0–13.0 and >13.0 minutes, respectively. cfDNA of pregnant women bearing a male fetus was similarly fractionated. qPCRs of the cfDNA fractions were performed for leptin (LEP) and SRY. LEP gene was detected in the fractions of short and long cfDNA, and thus, is present in both maternal and fetal alleles. In contrast to LEP, SRY was only found in fractions 1 and 2, corresponding to cfDNA fragments smaller than 500 bp and to fetal cfDNA (36), confirming the successful size-separation of cfDNA in pregnant women using the microdevice (25).
Lab-on-a-chip (LOC) capillary electrophoresis
A few years ago, a new technique, based on glass LOC capillary electrophoresis, has been introduced to separate genomic and synthetic DNA fragments of 20–40, 100, 200, 300 and 500 bp. Basically, the glass LOC comprises microfluidic channels and reservoirs enabling injection, separation and extraction of selected DNA fractions. The channels are filled with linear polymer sieving matrix, while the reservoirs are filled with standard running buffer (TBE). After introduction of a DNA sample into the reservoir, all subsequent steps of analysis are performed automatically by the docking station, a voltage control unit and a fluorometric detection unit for real-time monitoring of the DNA flow in the microfluidic channels. First, the fluorescent-labeled DNA sample is injected into the separation channel where DNA fragments become separated. Next, at the end of the separation channel, each DNA fragment is detected. Finally, the selected DNA fragment is moved to one of the two extraction channels by the automatic change in the separation voltage. DNA fragments of 20–500 bp can be successfully separated within 90 seconds, as documented by the fluorescence signals on the digital electropherograms.
This technique has potential of a fast and repeatable size-separation of cfDNA, i.e., fetal from maternal cfDNA, with a low risk of DNA contamination (26).
AmpFlSTR® Yfiler® PCR Amplification Kit
Kimura et al. investigated the size distribution of fetal cfDNA in maternal plasma of women using a commercial kit (AmpFlSTR® Yfiler® PCR Amplification Kit, Applied Biosystems, Foster City, CA, USA), which combines PCR and electrophoresis. This kit was originally designed with the intention to forensically recognize male perpetrators of crimes and permit the analysis of compound male-female DNA mixtures that often give equivocal or inconclusive results with autosomal short-tandem repeats (STR). The kit amplifies simultaneously 17 Y-chromosomal STR (Y-STR) loci in a single reaction. The subsequent performance of capillary electrophoresis with an allelic ladder marker documents the size of the obtained amplicons. Furthermore, the method in combination with sex determining region Y (SRY) primers for the amplicons of 193, 313, 392 and 524 bp allows determining the maximal size of fetal cfDNA fragments, which varied between 219 and 313 bp. Kimura et al. suggested that this size of cfDNA fragments may represent double nucleosomal complexes of ~146 bp (51).
PCR-based techniques
Real-time PCR
The amplification of amplicons of different sizes by real-time PCR (qPCR) can serve as a method to check DNA integrity. One of the first qPCR assays for measuring cfDNA integrity is the amplification of repeated sequences of ALU elements. An amplicon of 115 bp (ALU115) is usually amplified to measure the levels of truncated, apoptotic DNA and simultaneously, a 247-bp amplicon to measure longer DNA fragments. The data obtained from both DNA fragment lengths are compiled to calculate the integrity index of cfDNA (52). To date, the integrity index of cfDNA has been estimated by amounts of long versus short PCR products, and calculated in many tumor types, such as bladder (53), prostate (54), ovarian (55) and breast cancer (5).
Using the qPCR method, it has also been shown that nasopharyngeal carcinoma patients have a higher number of longer cfDNA molecules than healthy controls in their blood (16). On contrary, other studies showed that DNA released from malignant cells of nude athymic mice xenografted with HT29 cells and CRC patients is highly fragmented in comparison with DNA originating from normal tissues. Moreover, in the same study the ctDNA integrity of 133- vs. 290-bp fragments of the human ß-actin gene from xenografted mice pointed to an increase in fragmentation of ctDNA in line with the tumor weight and concentration of ctDNA. The same researchers also found that the efficiency of quantification of ctDNA by qPCR is influenced by the length of the amplicon. They showed that amplicons of a size between 60- and 100-bp have the most appropriate length for qPCR (22).
PCR-based size-selective enrichment of short cfDNA
Recently, a new PCR-based technique for size-selective enrichment and overall-amplification of short cfDNA fragments was established. It is an improved amplified fragment length polymorphism (AFLP) technique, in which cfDNA is (I) treated by T4 DNA polymerase to obtain blunt ends, (II) extended by adenine bases to create 3' sticky ends, (III) ligated with double stranded unidirectional linkers of 3'-thymine ends and (IV) simultaneously phosphorylated at the 5' end. The amplification of all cfDNA molecules in a single reaction system, independent of their particular sequences and lengths, can be carried out with a universal primer, since all cfDNA-linker molecules have the same sequence at the 5' end. Moreover, a DNA polymerase without 5' 3' exonuclease activity is used, and a modified PCR buffer is introduced to reduce the negative influence of GC content on PCR. The temperature of the denaturation step in the PCR cycle is decreased to suppress longer DNA fragments and allow only short DNA molecules to be amplified. This technique allows the selective enrichment of short cfDNA fragments. It was tested by the amplification of the SRY gene in plasma of pregnant women bearing a male fetus, and was proved to selectively size-enrich fetal cfDNA molecules (56). Thus, this method enriches short cfDNA fragments sufficient to meet the requirements of routine prenatal testing.
The above-described method has advantages as well as disadvantages. It can decrease the maternal cfDNA background, ameliorate the amplification and enrichment of fetal cfDNA, enhance the sensitivity of detection of fetal genetic abnormalities, decrease prenatal diagnosis costs and reduce special equipment demand. However, the method is time-consuming, consists of many laborious steps and includes repeated purification steps which may lead to the loss of DNA (56).
dPCR
dPCR relies on the random distribution of the template DNA in very small portions (droplets), containing only one or no copy of targeted DNA. By this technique, thousands of partitions can be created, in each of which a separated assay is carried out. In each sample portion, PCR is performed with a fluorescent probe, signals from all parallel positive reactions are digitally counted, and the proportion of positive and negative reactions is established. The copy number of the amplified targets is absolutely quantified, without the requirement of a standard curve. Since dPCR separates template DNA into partitions, it is much more sensitive than traditional qPCR, and even very low-abundant DNA sequences can be detected (38,41,57).
The dPCR of different amplicon lengths (49–309 bp) was recently used to analyze the abundance and fragment length distribution of mcfDNA in the plasma of lung transplant recipients. This study showed that mcfDNA fragmentation is increased in comparison with nuclear genomic DNA. This size-selective measurement showed that mcfDNA is much more abundant in the plasma of transplant recipients as previously assumed (15). The mcfDNA concentrations were 56 times higher than those of nuclear DNA (42).
To sum up, PCR-based techniques are very sensitive, but also limited by different amplification efficiencies, reproducibility and amplification errors (58).
MPS
MPS allows simultaneous sequencing of millions of cfDNA fragments. By the use of paired-end sequencing variants, each cfDNA molecule is read from both ends. The obtained sequences are, then, aligned to a reference genome (12,15,29,59). This accurate measurement of the length of each single cfDNA fragment is based on the determination of the distance between start and end coordinates of the paired-end reads. Length measurement of each cfDNA molecule is feasible, and the size profiles of cfDNA can be analyzed at single-base resolution in a genome-wide scale. The relative amounts of cfDNA of different lengths can be calculated with high precision (13,15,17,18). MPS size-based analyses of cfDNA were used, among the others, in prenatal diagnostics, to detect sub-chromosomal CNAs in maternal and fetal cfDNA (13), different types of fetal aneuploidies (12) and chromosomal deletions and duplications (39); in cancer research to assess mutant allele frequency in different cfDNA size fractions (34), hepatocellular carcinoma sequences and tumor-associated CNAs (15); in transplant recipients to study post-transplantation chimerism and origin of cfDNA; and in SLE to examine cfDNA abnormalities and their potential influence on the course of the disease (18).
Optimized MPS protocols
A typical MPS library preparation protocol consists of following steps: (I) end repair; (II) adenine tailing; (III) adaptor ligation; (IV) library amplification, and a purification step after each step. Standard MPS is usually based on a ligation of dsDNA sequencing adapters to the cfDNA molecules and a size-based removal of unwanted adapter dimers. However, this method is not effective for highly degraded, partially single-stranded DNA or very small cfDNA molecules, such as bacterial DNA or mcfDNA, and results in missing many cfDNA molecules. A recently introduced MPS method, which is based on single-stranded ligation and excludes a bead-based step of elimination of shorter cfDNA fragments, has improved the sensitivity to the different cfDNA species. The high efficiency of this method to detect, quantify and determine fragment length profiles of nuclear, microbial and mitochondrial cfDNA was among others demonstrated by the fact that the proportion of sequenced nuclear cfDNA fragments shorter than 100 bp in the pool of nuclear cfDNA was significantly increased to more than 20% (42).
Subsequently, MPS was further improved for the use of mcfDNA by combining end-repair and adenine tailing in a single reaction that is directly followed by adaptor ligation. This procedure avoids several purification steps that lead to the loss of short cfDNA fragments. Besides, mcfDNA is enriched prior to sequencing by a size-selection of 150–190 bp library fragments (estimated mcfDNA size range plus length of the adaptor) using automated DNA gel electrophoresis and size-based purification. The functionality of these modifications was confirmed by the improved isolation of short mcfDNA fragments and the subsequent detection of heteroplasmy. The proportion of mcfDNA fragments shorter than 100 bp was 19% using this improved protocol and was significantly higher than using the standard MPS (less than 2%) (19).
Microfluidic single molecule analysis
A method to quantify and measure the size of fluorescently labeled cfDNA directly from human serum has also been developed. Here, a combination of microfluidic cylindrical illumination confocal spectroscopy (µCICS) and fluorescent burst size analysis are applied. A special design of the µCICS platform ensures 100% detection efficiency of cfDNA molecules flowing through this construction, and provides fluorescent burst size dependent on the DNA length. Only a single reagent, a DNA intercalating dye (TOTO-3), is utilized in the assay. The number of TOTO-3 molecules incorporating in cfDNA is directly proportional to the DNA length. This dye was, therefore, chosen, because (I) it is highly specific to DNA, (II) it has a high fluorescent enhancement, to detect serum cfDNA, (III) its fluorescence can be distinguished from cellular auto-fluorescence, and (IV) it does not pass through cellular membranes, thereby preventing from the intercalation into nuclear DNA.
To verify the utility of this device, a 3-step experiment was performed. First, enzymatically digested lambda DNA was used to prepare the size calibration curve, that fluorescent burst adequately increased to the length of measured DNA. Next, lambda DNA was randomly fragmented by sonication during different time durations. After labeling, the length of lambda DNA fragments was measured by a microfluidic device. The calculated DNA fragmentation increased with the duration of sonication treatment. This assessment warranted to determine the integrity of serum cfDNA of early and late stage lung cancer patients. In this study, cfDNA was labeled directly in the serum, quantified and size-analyzed. Lung cancer patients at stage IV had a significantly higher proportion of longer cfDNA fragments than patients at stage I. The largest difference was observed in cfDNA fragments of ~800 bp, where patients at stage IV had almost 7 times higher DNA yields than patients at stage I.
To sum up, the present method may be an interesting alternative to PCR-based methods, since it allows a rapid, facile and inexpensive single molecule analysis of cfDNA integrity. By excluding the cfDNA isolation step, a potentially experimental bias can be eliminated (60).
Size-selective separation by lysine-functionalized silica particles
Recently, a new technique of size-dependent separation of DNA by lysine functionalized silica particles (Lys-SiO2) has been introduced. Although the silica matrix itself is known to adsorb DNA molecules on its surface by building “salt-bridges” in the presence of salt molecules in its environment, the adsorption of short DNA fragments by this phenomenon is poor. Even a modification of silica matrix with metal ions or amine groups did not solve this issue. To overcome this problem, lysine molecules were functionalized on the surface of silica particles. Due to this alteration, silica particles became sensitive to pH changes, and consequently, at- and detachment of DNA fragments on their surface can be easily controlled by changes of the environmental pH. Since the electrostatic attraction of DNA molecules to positively charged surface depends on their length, and thus, their total negative charge, the binding of DNA fragments of different lengths at Lys-SiO2 can be readily manipulated by regulating ionic strength. Higher concentrations of salt increase the distance between the charges, and the influence of this phenomenon rises along with the decrease in DNA length. By the adjustment of pH and salt concentration together, Lys-SiO2 allows an efficient separation of DNA fragments of different lengths. DNA of 101 bp length was demonstrated to be effectively isolated from DNA molecules of 1,073, 745 and 408 bp length (61).
The size fractionation with Lys-SiO2 can be applied in prenatal and cancer diagnosis, but also may serve as a tool for library preparation prior to MPS.
PME DNA Sizing Kit
PME DNA Sizing Kit (Analytik Jena, Jena, Germany) combines the isolation and the subsequent size-fractionation of cfDNA. In the first step, cfDNA is enriched by a polymer and afterwards cfDNA is extracted with the use of a spin column-based method. Finally, the eluate containing isolated cfDNA can be size-fractionated. Longer cfDNA fragments are bound at the first spin column. The size section of these cfDNA molecules is determined by adding appropriate amounts of binding conditioner solution, which has to be established empirically for the desired cfDNA size cutoff. cfDNA fragments shorter than the cutoff are released in the flow-through, and can be recovered on the second spin column. An advantage of this kit is that cfDNA can be enriched from a wide range of sample types (e.g., plasma, serum, urine) and volumes.
The PME DNA Sizing Kit was used to compare the cfDNA lengths in women suffering from intrahepatic cholestasis of pregnancy (ICP) with those in healthy pregnant women. Significantly higher proportions of cfDNA shorter than 200 bp were detected in women with ICP than in healthy controls (up to 3-fold difference). In addition, the copy number of cfDNA in the fraction longer than 200 bp was comparable between both cohorts. These results show that women with ICP have higher levels of plasma DNA of nuclear origin and higher cfDNA fragmentation than healthy controls (62).
Conclusions
The size distribution of cfDNA has been demonstrated to vary under different pathological and physiological conditions. In healthy and diseased (benign and malignant) individuals, the release processes of DNA into the human blood circulation, such as apoptosis, necrosis and active secretion, as well as the extent of cfDNA degradation by DNases in blood, differ resulting in a broad repertoire of different lengths of cfDNA between both groups. This varying prevalence permits a size-based separation of cfDNA that may contribute to improve the sensitivity of analytical applications. The use of size-fractionated cfDNA that reflects largely its origin also allows a better detection of genetic abnormalities. In particular, this may facilitate investigations of ctDNA in the plasma/serum of cancer patients, to trace the course of their disease and therapy success. To date, clinical relevance and application of these enquiries have only been found in fetal medicine. Accordingly, the different lengths of fetal and maternal cfDNA in maternal plasma have been shown to be useful for the prenatal diagnosis of certain fetal disorders.
In this study, we introduced numerous different methods to size-fractionate cfDNA (Table 2). Partly, these techniques showed no congruent cfDNA size patterns within the “same” patient populations. This discrepancy pointed among others to the different specificity of the techniques, different sample types (i.e., plasma, serum, urine) and small analyzed patient populations with different clinical parameters and risk factors. Therefore, selection and validation of the techniques using large and well-designed patient cohorts with similar parameters should be done, to obtain comparable results and standardize the most appropriate method for its clinical introduction.

Full table
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research for the series “Technologies in Liquid Biopsies - Potential applications in Medicine”. The article has undergone external peer review.
Conflicts of Interest: T Hillebrand is cofounder and CEO of AJ Innuscreen GmbH, and coinventor of patent applications describing method of size separation of cfDNA. M Grunt is employed by AJ Innuscreen GmbH. H Schwarzenbach has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mandel P, Metais P. Les acides nucleiques du plasma sanguin chez l’homme. C R Seances Soc Biol Fil 1948;142:241-3. [PubMed]
- Volik S, Alcaide M, Morin RD, et al. Cell-free DNA (cfDNA): Clinical Significance and Utility in Cancer Shaped By Emerging Technologies. Mol Cancer Res 2016;14:898-908. [Crossref] [PubMed]
- Salvi S, Gurioli G, De Giorgi U, et al. Cell-free DNA as a diagnostic marker for cancer: current insights. Onco Targets Ther 2016;9:6549-59. [Crossref] [PubMed]
- Gahan PB, Anker P, Stroun M. Metabolic DNA as the origin of spontaneously released DNA? Ann N Y Acad Sci 2008;1137:7-17. [Crossref] [PubMed]
- Umetani N, Giuliano AE, Hiramatsu SH, et al. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol 2006;24:4270-6. [Crossref] [PubMed]
- Wei F, Lin CC, Joon A, et al. Noninvasive saliva-based EGFR gene mutation detection in patients with lung cancer. Am J Respir Crit Care Med 2014;190:1117-26. [Crossref] [PubMed]
- Soltani M, Nemati M, Maralani M, et al. Cell-free fetal DNA in amniotic fluid supernatant for prenatal diagnosis. Cell Mol Biol (Noisy-le-grand) 2016;62:14-7. [PubMed]
- Tsui NB, Jiang P, Chow KC, et al. High Resolution Size Analysis of Fetal DNA in the Urine of Pregnant Women by Paired-End Massively Parallel Sequencing. PloS One 2012;7:e48319 [Crossref] [PubMed]
- Salvi S, Gurioli G, Martignano F, et al. Urine Cell-Free DNA Integrity Analysis for Early Detection of Prostate Cancer Patients. Dis Markers 2015;2015:574120 [PubMed]
- Lo YM, Patel P, Wainscoat JS, et al. Prenatal sex determination by DNA amplification from maternal peripheral blood. Lancet 1989;2:1363-5. [Crossref] [PubMed]
- Kagan KO, Sonek J, Wagner P, et al. Principles of first trimester screening in the age of non-invasive prenatal diagnosis: screening for chromosomal abnormalities. Arch Gynecol Obstet 2017; [Epub ahead of print]. [PubMed]
- Yu SC, Chan KC, Zheng YW, et al. Size-based molecular diagnostics using plasma DNA for noninvasive prenatal testing. Proc Natl Acad Sci U S A 2014;111:8583-8. [Crossref] [PubMed]
- Yu SC, Jiang P, Chan KC, et al. Combined Count- and Size-Based Analysis of Maternal Plasma DNA for Noninvasive Prenatal Detection of Fetal Subchromosomal Aberrations Facilitates Elucidation of the Fetal and/or Maternal Origin of the Aberrations. Clin Chem 2017;63:495-502. [Crossref] [PubMed]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. [Crossref] [PubMed]
- Jiang P, Chan CW, Chan KC, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A 2015;112:E1317-25. [Crossref] [PubMed]
- Chan KC, Leung SF, Yeung SW, et al. Persistent aberrations in circulating DNA integrity after radiotherapy are associated with poor prognosis in nasopharyngeal carcinoma patients. Clin Cancer Res 2008;14:4141-5. [Crossref] [PubMed]
- Lo YM, Chan KC, Sun H, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med 2010;2:61ra91 [Crossref] [PubMed]
- Zheng YW, Chan KC, Sun H, et al. Nonhematopoietically derived DNA is shorter than hematopoietically derived DNA in plasma: a transplantation model. Clin Chem 2012;58:549-58. [Crossref] [PubMed]
- Zhang R, Nakahira K, Guo X, et al. Very Short Mitochondrial DNA Fragments and Heteroplasmy in Human Plasma. Sci Rep 2016;6:36097. [Crossref] [PubMed]
- Schwarzenbach H, Pantel K. Circulating DNA as biomarker in breast cancer. Breast Cancer Res 2015;17:136. [Crossref] [PubMed]
- Thierry AR, El Messaoudi S, Gahan PB, et al. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev 2016;35:347-76. [Crossref] [PubMed]
- Mouliere F, Robert B, Arnau Peyrotte E, et al. High Fragmentation Characterizes Tumour-Derived Circulating DNA. PLoS One 2011;6:e23418 [Crossref] [PubMed]
- Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Chandrananda D, Thorne NP, Bahlo M. High-resolution characterization of sequence signatures due to non-random cleavage of cell-free DNA. BMC Med Genomics 2015;8:29. [Crossref] [PubMed]
- Hahn T, Drese KS, O’Sullivan CK. Microsystem for Isolation of Fetal DNA from Maternal Plasma by Preparative Size Separation. Clin Chem 2009;55:2144-52. [Crossref] [PubMed]
- Kubicki W, Walczak R. Preliminary Studies on Cell-free Fetal DNA Separation and Extraction in Glass Lab-on-a-chip for Capillary Gel Electrophoresis. Procedia Eng 2012;47:1315-8. [Crossref]
- Chiu RW, Akolekar R, Zheng YW, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ 2011;342:c7401. [Crossref] [PubMed]
- Mittra I, Nair NK, Mishra PK. Nucleic acids in circulation: are they harmful to the host? J Biosci 2012;37:301-12. [Crossref] [PubMed]
- Chan RW, Jiang P, Peng X, et al. Plasma DNA aberrations in systemic lupus erythematosus revealed by genomic and methylomic sequencing. Proc Natl Acad Sci U S A 2014;111:E5302-11. [Crossref] [PubMed]
- Enninga EA, Nevala WK, Holtan SG, et al. Immune Reactivation by Cell-Free Fetal DNA in Healthy Pregnancies Re-Purposed to Target Tumors: Novel Checkpoint Inhibition in Cancer Therapeutics. Front Immunol 2015;6:424. [Crossref] [PubMed]
- Anker P, Stroun M, Maurice PA. Spontaneous release of DNA by human blood lymphocytes as shown in an in vitro system. Cancer Res 1975;35:2375-82. [PubMed]
- Gahan PB. Circulating Nucleic Acids in Early Diagnosis, Prognosis and Treatment Monitoring: An Introduction. Springer, 2014:476.
- Pisetsky DS. Anti-DNA antibodies--quintessential biomarkers of SLE. Nat Rev Rheumatol 2016;12:102-10. [Crossref] [PubMed]
- Underhill HR, Kitzman JO, Hellwig S, et al. Fragment Length of Circulating Tumor DNA. PLOS Genet 2016;12:e1006162 [Crossref] [PubMed]
- Li Y, Zimmermann B, Rusterholz C, et al. Size separation of circulatory DNA in maternal plasma permits ready detection of fetal DNA polymorphisms. Clin Chem 2004;50:1002-11. [Crossref] [PubMed]
- Chan KC, Zhang J, Hui AB, et al. Size distributions of maternal and fetal DNA in maternal plasma. Clin Chem 2004;50:88-92. [Crossref] [PubMed]
- Dar P, Curnow KJ, Gross SJ, et al. Clinical experience and follow-up with large scale single-nucleotide polymorphism-based noninvasive prenatal aneuploidy testing. Am J Obstet Gynecol 2014;211:527.e1-e17. [Crossref] [PubMed]
- Lun FM, Chiu RW, Chan KC, et al. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin Chem 2008;54:1664-72. [Crossref] [PubMed]
- Yin AH, Peng CF, Zhao X, et al. Noninvasive detection of fetal subchromosomal abnormalities by semiconductor sequencing of maternal plasma DNA. Proc Natl Acad Sci U S A 2015;112:14670-5. [Crossref] [PubMed]
- Rumore PM, Steinman CR. Endogenous circulating DNA in systemic lupus erythematosus. Occurrence as multimeric complexes bound to histone. J Clin Invest 1990;86:69-74. [Crossref] [PubMed]
- Kisat MT. Circulating Mitochondrial and Bacterial DNA as Biomarkers of Sepsis. 2016 Jan 1 [cited 2017 Aug 24]. Available online: http://arizona.openrepository.com/arizona/handle/10150/621839
- Burnham P, Kim MS, Agbor-Enoh S, et al. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci Rep 2016;6:27859. [Crossref] [PubMed]
- Ellinger J, Albers P, Müller SC, et al. Circulating mitochondrial DNA in the serum of patients with testicular germ cell cancer as a novel noninvasive diagnostic biomarker. BJU Int 2009;104:48-52. [Crossref] [PubMed]
- Gordon PM, Khan A, Sajid U, et al. An Algorithm Measuring Donor Cell-Free DNA in Plasma of Cellular and Solid Organ Transplant Recipients That Does Not Require Donor or Recipient Genotyping. Front Cardiovasc Med 2016;3:33. [Crossref] [PubMed]
- Rohland N, Reich D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res 2012;22:939-46. [Crossref] [PubMed]
- Mouliere F, Piskorz AM, Chandrananda D, et al. Selecting Short DNA Fragments In Plasma Improves Detection Of Circulating Tumour DNA. bioRxiv 2017; [Crossref]
- Jorgez CJ, Bischoff FZ. Improving Enrichment of Circulating Fetal DNA for Genetic Testing: Size Fractionation Followed by Whole Gene Amplification. Fetal Diagn Ther 2009;25:314-9. [Crossref] [PubMed]
- Bronkhorst AJ, Wentzel JF, Aucamp J, et al. Characterization of the cell-free DNA released by cultured cancer cells. Biochim Biophys Acta 2016;1863:157-65. [Crossref] [PubMed]
- Heitzer E, Auer M, Hoffmann EM, et al. Establishment of tumor-specific copy number alterations from plasma DNA of patients with cancer. Int J Cancer 2013;133:346-56. [Crossref] [PubMed]
- Heidary M, Auer M, Ulz P, et al. The dynamic range of circulating tumor DNA in metastatic breast cancer. Breast Cancer Res 2014;16:421. [Crossref] [PubMed]
- Kimura M, Hara M, Itakura A, et al. fragment size analysis of free fetal DNA in maternal plasma using Y-STR loci and SRY gene amplification. Nagoya J Med Sci 2011;73:129-35. [PubMed]
- Umetani N, Kim J, Hiramatsu S, et al. Increased integrity of free circulating DNA in sera of patients with colorectal or periampullary cancer: direct quantitative PCR for ALU repeats. Clin Chem 2006;52:1062-9. [Crossref] [PubMed]
- Ellinger J, Bastian PJ, Ellinger N, et al. Apoptotic DNA fragments in serum of patients with muscle invasive bladder cancer: a prognostic entity. Cancer Lett 2008;264:274-80. [Crossref] [PubMed]
- Ellinger J, Bastian PJ, Haan KI, et al. Noncancerous PTGS2 DNA fragments of apoptotic origin in sera of prostate cancer patients qualify as diagnostic and prognostic indicators. Int J Cancer 2008;122:138-43. [Crossref] [PubMed]
- Salani R, Davidson B, Fiegl M, et al. Measurement of Cyclin E Genomic Copy Number and Strand Length in Cell-Free DNA Distinguish Malignant versus Benign Effusions. Clin Cancer Res 2007;13:5805-9. [Crossref] [PubMed]
- Yang Q, Du Z, Song Y, et al. Size-selective separation and overall-amplification of cell-free fetal DNA fragments using PCR-based enrichment. Sci Rep 2017;7:40936. [Crossref] [PubMed]
- Hayden RT, Gu Z, Ingersoll J, et al. Comparison of Droplet Digital PCR to Real-Time PCR for Quantitative Detection of Cytomegalovirus. J Clin Microbiol 2013;51:540-6. [Crossref] [PubMed]
- Diehl F, Diaz LA. Digital quantification of mutant DNA in cancer patients. Curr Opin Oncol 2007;19:36-42. [Crossref] [PubMed]
- Wong FCK, Sun K, Jiang P, et al. Cell-free DNA in maternal plasma and serum: A comparison of quantity, quality and tissue origin using genomic and epigenomic approaches. Clin Biochem 2016;49:1379-86. [Crossref] [PubMed]
- Liu KJ, Wang TH. PCR-free, microfluidic single molecule analysis of circulating nucleic acids in lung cancer patient serum. Conf Proc IEEE Eng Med Biol Soc 2011;2011:8392-5. [PubMed]
- Liu L, Guo Z, Huang Z, et al. Size-selective separation of DNA fragments by using lysine-functionalized silica particles. Sci Rep 2016;6:22029. [Crossref] [PubMed]
- Vlková B, Kalousová M, Germanová A, et al. Cell-free DNA is higher and more fragmented in intrahepatic cholestasis of pregnancy. Prenat Diagn 2016;36:1156-8. [Crossref] [PubMed]


