
Detection of microRNAs in circulating tumor cells
Introduction
During recent years, emerging efforts have been directed towards establishing valuable biomarkers for clinical diagnostics, prediction of treatment response and estimation of prognosis in cancer patients. Among those that have gained acceptance as prognostic and potential predictive biomarkers are circulating tumor cells (CTCs) detectable in peripheral blood as liquid biopsy (1). Although only in a minority of patients with early stage cancer CTCs are found, CTC detection in the context of adjuvant or neoadjuvant therapy, e.g., in breast cancer, is associated with clinical outcome (2,3). Moreover, in the metastatic disease setting CTCs can support therapeutic monitoring and stratification of patients for an individual cancer therapy (4). Various techniques to enrich and detect CTCs from whole blood or peripheral blood mononuclear cells (PBMCs) are readily available yet, however, only one approach, the CellSearch® system (Menarini-Silicon Biosystems, Bologna, Italy, formerly Janssen Diagnostics), assures CTC detection in a standardized assay format. This system allows the detection of epithelial cancer cells expressing epithelial cell adhesion molecule (EpCAM) and keratin in an automated fashion and has been cleared by the Food and Drug Administration (FDA) for the analysis of blood samples from patients with metastatic breast, colorectal and prostate cancer (5-7). Since the majority of currently available methods for CTC detection is based on epithelial cell features as surrogate markers for epithelial tumor cells, finding CTCs that lost epithelial and gained mesenchymal characteristics is challenging. Other platforms applied to trace also mesenchymal-like CTCs have been established, but still lack specificity and broad-range clinical validation (8). To complement CTC enumeration, several down-stream analyses have been developed to isolate and characterize CTCs at proteomic, genomic, transcriptomic and epigenetic level including single cell analyses (9). This has not only improved our current knowledge about tumor cell dissemination and minimal residual disease (MRD), but has also contributed to the identification of therapeutically relevant targets on CTCs, such as human epidermal growth factor receptor-2 (HER2), estrogen receptor (ER), or prostate specific membrane antigen (PSMA), predominantly in advanced tumor stages where tumor tissues are only rarely available (10-13). Moreover, first interventional clinical studies based on CTC detection and characterization have been initiated (14,15).
A multitude of genetic and epigenetic alterations in tumor suppressor genes and/or oncogenes contributes to malignant transformation of normal to tumor cells which are characterized e.g., by increased proliferation, migratory and invasive ability. Importantly, some tumor cells are capable to disseminate from the primary tumor, to resist shear forces and immune attacks in the circulation, and eventually to initiate metastases at distant sites. Moreover, subsets of tumor cells may also acquire features rendering them resistant to cancer therapies. These different tumor characteristics are caused by various disorders in the complex regulatory network of signaling pathways ensuring balanced growth, differentiation, senescence and cell death under physiological conditions. There is growing evidence that miRNAs which are suggested to be involved in the complex regulation of gene expression in cancer cells play a crucial role in initiation and progression of cancer by interfering with cell cycle control, DNA damage response, apoptosis, differentiation, autophagy, epithelial to mesenchymal transition (EMT) and metastasis as hallmarks of cancer (16,17).
Compared to the already discovered importance of miRNAs in tumor tissue and of circulating cell-free miRNAs in plasma or serum, less is known about the expression of miRNAs in CTCs. Moreover, clinical studies including miRNA analyses of CTCs are still in their infancy. Therefore, this article summarizes existing efforts to detect and characterize miRNAs in CTCs present in peripheral blood of carcinoma patients. Furthermore, the potential of CTC miRNA expression to add value complementary to CTC enumeration and quantification is discussed.
Impact of microRNAs on tumorigenesis
MiRNAs are short non-coding RNAs of 18–25 nucleotides in length which bind to the 3'UTR (untranslated region) of mRNAs to inactivate gene expression. However, miRNAs are also able to bind to the 5'UTR region and to open reading frames (ORFs) to activate gene expression (18-20). Transcription of genes encoding miRNAs is usually performed by RNA polymerase II generating primary miRNAs. After cleaving of these miRNAs by RNase III Drosha, attained precursor miRNAs (70 to 100 nucleotides in length) are transferred from the nucleus into the cytoplasm for further cleavage by RNase III Dicer into mature double-stranded RNAs. These RNAs are now loaded into the miRNA-associated RNA induced silencing complex (RISC). Here, they work as post-transcriptional regulators and add a further layer in the complex regulation of tumor-associated genes and proteins (19,20). Both genetic and epigenetic aberrations in miRNA genes and their regulatory regions as well as their location in fragile chromosomal regions might contribute to the dysregulation of miRNA expression in cancer cells (16,21).
MiRNAs can either degrade mRNAs or repress translation dependent on the degree and nature of complementary sites between the miRNA and the target mRNA (22). Moreover, they can directly or indirectly influence the expression of both oncogenes and tumor suppressor genes (23). To this end, dysregulation of expression and function of miRNAs has been described for almost all tumor entities. MiRNAs can be classified as oncogenic by down-regulating the expression of tumor suppressor genes or as tumor-suppressive by down-regulating oncogenes (23). They are designated oncogenic or oncomiRs if they have a major role in initiation and/or progression of cancer. Moreover, involvement of miRNAs in modulating tumor-modifying extrinsic factors important for cancer progression and metastasis, such as interactions of cancer cells with the immune system or with stromal cells has been recently reported (23). These results suggest that either activation or suppression of the expression of specific miRNAs could be incorporated in currently established therapeutic strategies (24).
The number of known cancer-related miRNAs is continually increasing leaving function of most of them widely undefined so far. Besides their location in single chromosomal regions miRNA encoding genes can also be clustered on chromosomes and co-transcripted as polycistronic transcripts and thus coordinately regulate biological processes (25,26). Moreover, dysregulation of individual miRNAs might have profound effects on the expression of several hundred different mRNAs, thus hampering prediction of fundamental target mRNAs with key regulatory functions in cancer development and progression (27). So far, all known miRNAs are mapped and various new data platforms and bioinformatics tools enable prediction of novel miRNAs, miRNA networks, pathway-based approaches, functions and miRNA targets (19,24). Among others, the database miRBase (http://www.mirbase.org) and Kyoto Encyclopedia of Genes and Genomes (KEGG)-miRPath are valuable tools for miRNA expression and miRNA-associated signal transduction pathways, respectively (28-30). While miRs 17–92, 21, 10b, 106, 191 or 221/222, for instance belong to comprehensively validated oncogenic miRNAs in different cancer entities, other miRNAs such as let-7, miR-15/16, -200, or -141 rather belong to miRNAs with tumor-suppressive properties. Several recent review articles highlight the impact and function of miRNAs on different aspects of cancer initiation and metastasis (16,20,23,24,31,32). Results from a large number of studies on tissue samples suggest that miRNA profiles are valuable tools for distinguishing tumor from normal cells. Thus, the potential of miRNA expression in tumor tissue as diagnostic, prognostic and predictive biomarker has unambiguously been shown. Clinical usage, however, is mostly restricted to cases where primary tumor tissue is available while in the metastatic setting, tumor tissue is only accessible in selected cases (33). Consequently, miRNA analyses in liquid biopsies have gained attraction as diagnostically and therapeutically valuable approaches.
Methods for the detection of miRNAs
To date, there is no standardized method available to measure expression of individual miRNAs or miRNA profiles, leading to controversies in interpretation of miRNA results. Therefore, improving accuracy and reproducibility of assays aimed to detect miRNAs is of utmost importance. While in principle quantitative RT-PCR (qRT-PCR), microarrays or RNA sequencing (RNASeq) by Next generation sequencing (NGS) can be applied to determine tissue-associated miRNAs as well as circulating cell-free miRNAs in body fluids, in situ hybridization using locked nucleic acid (LNA) probes can only be used to detect cellular miRNA expression in cells or tissues (34-39). However, in situ hybridization has the advantage that heterogeneity of miRNA levels among tumor and stromal cells and cellular origins of miRNAs can be visualized. Nevertheless, qRT-PCR is the most straightforward method suitable for clinical routine. Especially TaqMan miRNA assays and miRNA-specific stem loop primers ensure highly specific detection of mature miRNAs. The different qRT-PCR approaches currently applied are summarized in recent review articles (34,40,41). For quantification of miRNA results, data normalization using endogenous and exogenous reference genes is required; however, thus far standardized methods for data correction are still lacking (42). Moreover, pre-analytical variables such as shipment of samples, storage, and RNA isolation as well as analytical performance criteria regarding sensitivity, reproducibility and specificity have to be defined and standardized to achieve comparability of results from different studies. Recommendations for best clinical practice, generation of workflows, aspects of automation and using emerging technologies for miRNA detection have been published recently (40,43).
Detection of circulating cell-free miRNAs
To date, rising evidence is provided that miRNAs can be released or secreted into extracellular compartments by a multitude of different cell types under physiological and pathological circumstances. Consequently, miRNAs are detectable in body fluids, such as plasma, serum or urine.
MiRNAs are characterized by high extracellular stability due to protection by being packaged in vesicles or by binding on Argonaute (AGO) or high-density lipoprotein (HDL) proteins. Hence, tumor-secreted extracellular vesicles (exosomes) can translocate miRNAs from cells of origin to recipient cells even at distant organs, and are therefore discussed now as cell-to-cell mediators of signals which are important e.g., for tumor cell spreading and metastasis (18,33,44,45). Results from numerous studies demonstrate that expression profiles of miRNAs in plasma and serum are associated with the clinical outcome of cancer patients (33,46). Although miRNAs detected in body fluids can potentially reflect the entire miRNA landscape in an individual tumor patient, to find the cellular origin of these miRNAs and to identify tumor-associated miRNAs, however, remains challenging. In this context it is important to know whether CTCs also release miRNAs and thereby contribute to detectable miRNA levels in body fluids derived from cancer patients. A correlation of the number of CTCs with levels of miRNAs in plasma collected from patients with metastatic breast cancer has already been reported (47,48).
Detection of miRNAs in CTCs
Compared to miRNAs in tumor tissue, plasma/serum or exosomes, less is known about the expression of miRNAs in CTCs. Since CTCs are rare cells they have to be pre-enriched from whole blood samples prior to detection and characterization. Several approaches relying either on physical properties of CTCs, such as density, size, plasticity or electrophoretic mobility or on the expression of antigens allowing capturing of CTCs, e.g., by antibodies coated to magnetic particles, have been established (8). Nevertheless, exploration of miRNAs in CTCs and comparison of results from different studies are hindered by (I) the very low concentration of these cells especially in early stage cancer; (II) divergent and not readily predictable numbers of contaminating leukocytes; (III) CTCs eluding common detection methods, e.g., by loss of epithelial cell features in the course of EMT and; (IV) the difficulty to select the best suited method to detect miRNAs compatible with a particular CTC enrichment and isolation method.
Detection of miRNA expression in blood samples enriched for PBMCs and CTCs
Circulating or disseminated tumor cells can be co-enriched with PBMCs by common blood cell separation methods such as Ficoll-Hypaque density gradient centrifugation and subsequently characterized by a multitude of immunological or molecular techniques (2,49,50). Although miRNA profiles of PBMCs including expression of oncomiRs such as miR-21 can be affected by different inherent non-cancerous conditions, inflammatory diseases, exercising, aging or smoking (51-55), there are several studies investigating the potential of miRNA expression in PBMCs as diagnostic marker for cancer patients (56). Thus, Zheng et al. detected higher miR-21 expression levels in PBMCs from patients with gastric cancer compared to those enriched from healthy persons and postulated that monitoring miR-21 in PBMCs might become a novel method to detect CTCs (57). In another study, miR-21 expression of PBMCs was increased in patients with prostate cancer compared to those suffering from benign prostate hyperplasia or in healthy controls and was associated with the survival of prostate cancer patients (58). Interestingly, NGS on PBMCs enriched from women at high risk for breast cancer unaffected at time of sample collection, revealed differences between miRNA profiles of women without breast cancer during follow up time and those later diagnosed with breast cancer. This led to conclude that miRNAs might become potential non-invasive biomarkers for breast cancer prediction (59). Interestingly, miRNAs in PBMCs could be identified as promising tools to differentiate between pancreatic cancer and benign/peri-pancreatic disease (60). Additionally, diagnostically relevant miRNA profiles assessed on PBMCs were identified for patients with non-small cell lung cancer (NSCLC) (61,62). However, these miRNA profiles did only have weak similarity with those identified in lung tumor tissues or in plasma/serum from NSCLC patients suggesting that these miRNAs are not likely to be originated from CTCs (61). Instead, dysregulation of miRNA expression of PBMCs might be indicative for example of early events in cancer immunogenicity or immune evasion (61). Unfortunately, simultaneous proof of CTCs was missing in all these studies.
To circumvent this disadvantage and to proof if miRNAs might be derived from CTCs, first pilot studies aimed to detect miRNA expression in blood samples further enriched for CTCs have been performed. However, no standard assay is available thus far to get rid of a wide range of normal blood cells. Assuming that the majority of CTCs originating from epithelial tumors is EpCAM-positive, capturing of CTCs from PBMCs by anti-EpCAM antibody-coated magnetic beads is an effective method to eliminate large proportions of leukocytes. Hence, Markou et al. selected miR-21, miR-146a, miR-200c and miR-210 known to be involved in the regulation of migration, invasion and metastasis of breast cancer and analyzed their expression in EpCAM-enriched CTCs (Figure 1A) and in corresponding plasma. Samples were collected from 55 patients with metastatic breast cancer as well as from 20 healthy controls and miRNA analysis was performed by qRT-PCR TaqMan microRNA assays (63). For the first time, miR-21 was reported to be up-regulated both in plasma as well as in EpCAM-enriched CTCs from the same patients. Interestingly, in plasma samples and corresponding EpCAM-enriched CTCs, all 4 miRNAs were overexpressed compared to the appropriate control samples from healthy individuals. In contrast, miRs-200c and -210 were down-regulated in primary tumor tissues compared to non-tumoral tissues in this study (63). Moreover, several studies demonstrated that overexpression of members of the miRNA-200 family is rather associated with suppression of EMT and inhibition of tumor progression and metastasis (69-71). However, there are also studies providing evidence for increased plasma or serum levels of miRNA-200c in advanced and metastatic tumor stages associated with local recurrence, distant metastasis and poor prognosis (72-76). In experimental systems, it was even demonstrated that miR-200 family members secreted in extracellular vesicles from metastatic murine and human breast cancer cell lines were able to transfer metastatic capacity and to promote metastasis of otherwise weakly metastatic cells by mediating MET (mesenchymal to epithelial transition) (77). Therefore, it is not surprising that significantly higher levels of circulating miRNA-200c were associated with CTC-positivity in metastatic breast cancer and with poor progression-free survival and metastasis up to 2 years prior to clinical diagnosis in breast cancer patients (78). Moreover, in another study on metastatic breast cancer, CTC-positive patients had significantly higher concentrations of circulating miR-200a-c and miR-210 than CTC-negative patients and controls, rendering these miRNAs promising candidates for prediction of prognosis and CTC status (47).
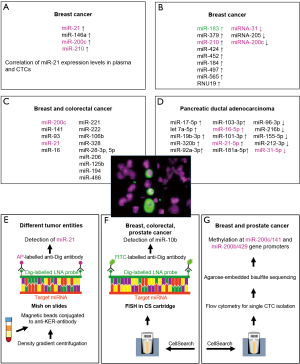
In the study presented by Sieuwerts et al. CTCs were enriched by anti-EpCAM antibody-coated ferrofluid using the CellSearch Profile Kit. Whole blood samples were collected from 50 patients with metastatic breast cancer before first line chemotherapy and from 53 healthy control persons. CTCs were enumerated in a sample simultaneously collected and processed with the CellSearch Epithelial Cell kit (48). For miRNA analysis, RNA was isolated, reverse-transcribed by a multiplex stem-loop approach and miRNAs were detected by multiplex and uniplex TaqMan qRT-PCR reactions. After screening of 436 miRNAs in tumor tissues obtained from 150 breast cancer patients, 39 miRNAs with at least 10-times higher expression in breast cancer tissues compared to CellSearch-enriched cells from 6 healthy blood donors were selected for further analyses in enriched CTCs. Furthermore, 4 miRNAs valuable for differentiation between ER-positive and –negative tumors were analyzed. Additionally, all miRNA data were validated in uniplex RT-PCR. Finally, 9 and 3 miRNAs were identified with significantly higher or lower levels of expression, respectively (Figure 1B) in blood samples from patients with ≥5 CTCs/7.5 mL compared to those obtained from healthy blood donors. Of note, only miR-183 could be considered as “CTC-specific”, since it was expressed at higher levels in blood samples from patients with ≥5 CTCs/7.5 mL (n=32, range 6–2,262) compared to those of patients with <5 CTCs (n=9). MiR-183 is frequently deregulated in numerous cancer entities (79) and has already been described as relevant for prognosis, regulation of proliferation and migration, inhibition of apoptosis, and maintenance of breast cancer stem cell phenotype (80-83). Sieuwerts et al. also analyzed mRNA profiles of enriched CTCs and detected 55 transcripts associated with ≥5 CTCs/7.5 mL compared to CellSearch enriched cells from healthy blood donors (48). Subsequent unsupervised 2-dimensional average linkage hierarchical cluster analysis using the 65-gene profile (55 miRNAs and 10 miRNAs with higher expression in patients with ≥5 CTCs than in healthy blood donors) resulted in a clustering of 4 groups of patients. Interestingly, the identified potentially CTC-associated miRNAs could be assigned to particular gene expression profiles. Thus, miR-183 for instance clustered to a subgroup of ER-negative CTCs suggesting that this miRNA might be negatively regulated by ER or is able to down-regulate ER (48).
Very recently, Leong and co-workers introduced a paper-based miRNA expression profiling starting with a cell sieve-assisted filtration method for EDTA blood to enrich CTCs (64). Subsequently identification of CTCs was performed by fluorescence staining with DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) to confirm nuclear integrity, by anti-EpCAM antibodies for proof of epithelial cell origin as surrogate for tumor cells and anti-CD45 antibodies to label leukocytes. Cells eluted from the microsieve, consisting of erythrocytes, leukocytes and CTCs were spotted onto a MicroWell MiniTray system thereby partitioning the eluted cells. Fluorescence microscopy-controlled CTC-containing wells were pooled together for miRNA extraction and purification using a cellulose paper-based Elute card chemically treated to immobilize nucleic acids from lysed cells (64). After multiplex stem-loop reverse transcription of extracted RNA, miRNA expression was analyzed by qRT-PCR. The authors selected miRNAs relevant for breast cancer metastasis or chemoresistance from literature searches and profiled cell sieve enriched cells from 21 breast or colorectal cancer patients and 3 healthy control persons. In this study, for 19 cancer patients, a more than 2.3-fold difference in the expression of at least one of selected miRNAs (Figure 1C) between CTC-containing and CTC-free samples was observed. Moreover, for one breast cancer patient, feasibility of high-throughput qPCR array analysis for CTC miRNA discovery was shown. Interestingly, longitudinal CTC-specific miRNA expression profiling of this patient was performed in the context of neoadjuvant chemotherapy. Here, different miRNA profiles in blood samples collected before and after chemotherapy were obtained with decreased miRNA-28-5p and miR-221 and increased miR-106b levels (64). Although this approach had the advantage that CTC-positivity could be controlled, the still preliminary results have to be validated in large prospective studies with clearly defined endpoints.
An ultra-specific microfluidic device combining inertial spiral microfluidics and EpCAM-immunomagnetic cell sorting for efficient isolation of CTCs from whole blood with ultra-high CTC purity and high throughput potential was recently reported by Jack and co-workers (65). Here, blood samples were processed from 14 patients with locally advanced or metastatic pancreatic ductal adenocarcinomas (PDAC) and 4 healthy individuals. Identification of CTCs occurred by immunofluorescence staining using antibodies against keratin-19 and CD45 as well as DAPI staining. CTC counts ranged between 14 and 938/mL in contrast to an average of 3 “CTC-like” cells/mL in healthy controls. After RNA isolation from the residual enriched cells, subsequent reverse transcription using the miRCURY LNA Universal RT-PCR kit, and qRT-PCR-arrays for profiling of 372 miRNAs were conducted. The proof-of-concept-device for miRNA profiling was proven on blood samples from two PDAC patients tested CTC-positive with 7 and 15 CTCs/mL, respectively, and from a healthy control. The authors claimed that the high purity of CTCs obtained by this device enables reliable miRNA detection in CTCs. Interestingly, among the miRNAs with high expression in CTCs were miR-210 and miR-21, which previously have been described as relevant for PDAC and other cancer entities (84-86). The 10 miRNAs with highest expression in CTCs are displayed in Figure 1D. In the study by Jack and co-workers, simultaneous mRNA analyses of CTCs revealed that miRNA expression might go along with characteristic mRNA profiles (65). Despite the low number of enrolled patients, the presented results demonstrate the potential of this workflow to analyze miRNA profiles on highly purified CTC preparations and pave the way for larger studies on well-defined cohorts of cancer patients.
Due to the lack of standardized methods, results of different studies analyzing miRNAs from enriched CTCs cannot reasonably be compared so far. This is not only due to different tumor entities and tumor stages, but also to strongly varying ratios between CTCs and co-enriched leukocytes and other blood cells, different absolute CTC numbers in the CTC-positive groups of patients and lack of information about EpCAM-negative CTCs. Thus, to reliably identify the cellular origin of miRNAs and to estimate heterogeneous distribution of miRNA expression, methods allowing investigation of single CTCs clearly distinguishable from blood cells have to be established.
Detection of miRNA expression in single CTCs
Thus far, only two reports about miRNA analysis in single CTCs performed by in situ hybridization are available (66,67).
Results of the first study that was aimed to analyze miR-21 expression at single CTC level using miRNA in situ hybridization (MiSH) were published by Ortega et al. (66). This miRNA was described as oncogenic miRNA in numerous studies (31,87) and is considered as very promising target of miRNA-directed therapeutic strategies (88). Since hematopoietic cells did not express detectable amounts of miR-21, the authors concluded that this miRNA is a very helpful diagnostic tool to trace minute amounts of CTCs (67). In this study, ISH applying digoxigenin-labeled LNA probes and alkaline phosphatase-labeled anti-digoxigenin antibodies, was conducted after enrichment of CTCs by Ficoll density gradient centrifugation and cell separation with magnetic microbeads conjugated to anti-keratin-antibodies (Figure 1E) (66). Detection of CTCs was simultaneously performed by immunofluorescence using fluorescein isothiocyanate (FITC)-labeled anti-keratin antibodies. Keratin-positive CTCs were detected in peripheral blood samples of 11/25 patients with metastatic cancer and miRNA-21-positive CTCs were identified in all cases.
In the second study presented by Gasch et al. (67), CTCs detected with the CellSearch system from 8, 1 and 2 patients suffering from metastatic breast, colorectal and prostate cancer, respectively were analyzed for miR-10b expression by FISH. Here, after CellSearch processing using the CellSearch Epithelial Cell kit (89), cartridges containing enriched CTCs were treated with methanol-glacetic acid. For miRNA detection in CTCs, a combined FISH and immunofluorescence staining was performed using digoxigenin-labeled LNA miRNA probes including control probes, FITC-labeled-digoxigenin-antibodies and phycoerythrin (PE)-and allophycocyanin (APC)-labeled keratin and CD45 antibodies, respectively, directly in the CellSearch cartridge (Figure 1F) (67). Fluorescence signals of 15 to 110 CTCs isolated from patient samples were evaluated microscopically. Interestingly, heterogeneous intensity of miRNA-10b expression was found among CTCs from individual patients with percentages of miR-10b-positive CTCs ranging between 20% and 100%. Several in vivo and in vitro studies demonstrate that miR-10b belongs to promising targets for miRNA-based cancer therapies (88,90) as it is associated with tumorigenesis and metastasis as well as with the prognosis of cancer patients (91,92). Moreover, miR-10b is designated as master regulator of metastatic cell viability (90).
Both studies using ISH to detect miRNAs in CTCs are proof-of-concept studies validated with appropriate controls, but involving only small numbers of patients. Moreover, ISH analyses are still restricted to selected panels of miRNAs. However, cellular origin and intensity of miRNA expression can be investigated, and modern microscopy and imaging technologies will open the way for the establishment of multi-miRNA testing also at single cell level.
MicroRNA expression itself can be regulated by different mechanisms including epigenetic alterations. For example, inactivation of miRNA encoding genes by methylation of their promoter regions has frequently been described (93). Thus, discovering methylation might permit meaningful information about expression levels of miRNA-encoding genes. Notably, Pixberg and co-workers recently established a method suitable even for single cell methylation analysis and thus applicable for CTCs (68). To do this, the authors developed a workflow for the analysis of CTCs detected with the CellSearch system, subsequent isolation of CTCs by flow cytometry sorting and methylation analysis of multiple loci by multiplex PCR, based on agarose-embedded bisulfite treatment (multiplexed scAEBS) (Figure 1G). The assay enabled simultaneous detection of promoter methylation of three EMT-associated genes (miR-200c/141, miR-200b/a/429 and E-Cadherin (CDH1) in single cells. CTCs and leukocytes were analyzed in parallel and methylation patterns of CTCs resembled those of epithelial-like cells. Blood samples from 11 and 6 patients with metastatic breast and prostate cancer, respectively, were investigated. Of note, individual CTCs exhibited specific methylation pattern suggesting that tumor-specific epigenetic regulation of EMT-associated genes is involved in tumor cell dissemination and spread. Interestingly, methylation of the promoter region of the miR-200 family genes was characterized by heterogeneous intensity among individual CTCs of patients with metastatic breast and prostate cancer (68). These important results pave the way for comprehensive epigenetic analyses of CTCs, which have the potential to complement proteomic, genomic and transcriptomic CTC analyses (9) in well-defined future clinical studies.
Conclusions
Results of first studies to detect miRNA expression in CTCs are promising to add value complementary to CTC enumeration and quantification. As CTCs inherently harbor information about the tumor in an appropriate stage, analysis of miRNAs in CTCs enables uncovering of regulatory mechanisms contributing to tumor development and metastasis. However, all published studies are proof-of-concept studies predominantly aimed to establish assay formats compatible with specific enrichment and detection methods of CTCs. Thus, numbers of patients enrolled are still low and validation in clinical studies is missing. Furthermore, CTCs are rare cells which have to be enriched with high sensitivity and specificity from blood samples prior to further investigations. Thus, miRNA analyses of enriched CTCs still face different problems such as (I) varying numbers of co-enriched contaminating leukocytes; (II) impact of different blood donors on miRNA expression in normal blood cells; (III) CTCs not detectable with current standard assays and; (IV) lack of information about tumor cell heterogeneity. In contrast, single CTC analyses enable visualization of tumor cell heterogeneity among individual CTCs, but are still restricted in the number of miRNAs that can simultaneously be tested.
In summary, deregulation of certain miRNAs, including among others miRs-16, -21, -31, -200 and -210 which are of paramount importance for maintenance of EMT/MET and metastatic progression was demonstrated in CTCs. Clinical significance of these results, however, remains to be elucidated on larger patient cohorts enrolled in well-defined clinical trials. The major challenge for future studies will be the assessment of standardized methods for sample collection and storage, enrichment and isolation of CTCs and miRNA analyses to ensure comparability of different studies and to shed more light on the potential of miRNAs in CTCs as blood-based biomarkers.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Heidi Schwarzenbach) for the series “Technologies in Liquid Biopsies - Potential applications in Medicine” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2017.10.24). The series “Technologies in Liquid Biopsies - Potential applications in Medicine” was commissioned by the editorial office without any funding or sponsorship. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Alix-Panabières C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov 2016;6:479-91. [Crossref] [PubMed]
- Rack B, Schindlbeck C, Juckstock J, et al. Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. J Natl Cancer Inst 2014;106:dju066 [Crossref] [PubMed]
- Riethdorf S, Muller V, Loibl S, et al. Prognostic impact of circulating tumor cells for breast cancer patients treated in the neoadjuvant "Geparquattro" trial. Clin Cancer Res 2017;23:5384-93. [Crossref] [PubMed]
- Pantel K, Alix-Panabieres C. Liquid biopsy: Potential and challenges. Mol Oncol 2016;10:371-3. [Crossref] [PubMed]
- Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91. [Crossref] [PubMed]
- Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:3213-21. [Crossref] [PubMed]
- de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9. [Crossref] [PubMed]
- Joosse SA, Gorges TM, Pantel K. Biology, detection, and clinical implications of circulating tumor cells. EMBO Mol Med 2015;7:1-11. [Crossref] [PubMed]
- Alix-Panabières C, Pantel K. Characterization of single circulating tumor cells. FEBS Lett 2017;591:2241-50. [Crossref] [PubMed]
- Riethdorf S, Muller V, Zhang L, et al. Detection and HER2 expression of circulating tumor cells: prospective monitoring in breast cancer patients treated in the neoadjuvant GeparQuattro trial. Clin Cancer Res 2010;16:2634-45. [Crossref] [PubMed]
- Babayan A, Hannemann J, Spotter J, et al. Heterogeneity of estrogen receptor expression in circulating tumor cells from metastatic breast cancer patients. PLoS One 2013;8:e75038 [Crossref] [PubMed]
- Gorges TM, Riethdorf S, von Ahsen O, et al. Heterogeneous PSMA expression on circulating tumor cells: a potential basis for stratification and monitoring of PSMA-directed therapies in prostate cancer. Oncotarget 2016;7:34930-41. [Crossref] [PubMed]
- Babayan A, Alawi M, Gormley M, et al. Comparative study of whole genome amplification and next generation sequencing performance of single cancer cells. Oncotarget 2016;8:56066-80. [PubMed]
- Schramm A, Friedl TW, Schochter F, et al. Therapeutic intervention based on circulating tumor cell phenotype in metastatic breast cancer: concept of the DETECT study program. Arch Gynecol Obstet 2016;293:271-81. [Crossref] [PubMed]
- Ignatiadis M, Rack B, Rothé E, et al. Liquid biopsy-based clinical research in early breast cancer: The EORTC 90091-10093 Treat CTC trial. Eur J Cancer 2016;63:97-104. [Crossref] [PubMed]
- Zhou K, Liu M, Cao Y. New insight into microRNA functions in cancer: oncogene-microRNA-Tumor Suppressor Gene Network. Front Mol Biosci 2017;4:46. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522-31. [Crossref] [PubMed]
- Shukla V, Varghese VK, Kabekkodu SP, et al. A compilation of Web-based research tools for miRNA analysis. Brief Funct Genomics 2017;16:249-73. [Crossref] [PubMed]
- Oliveto S, Mancino M, Manfrini N, et al. Role of microRNAs in translation regulation and cancer. World J Biol Chem 2017;8:45-56. [Crossref] [PubMed]
- Zare M, Bastami M, Solali S, et al. Aberrantly miRNA promoter methylation and EMT-involving miRNAs in breast cancer metastasis: diagnosis and therapeutic implications. J Cell Physiol 2018;233:3729-44. [Crossref] [PubMed]
- Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics 2010;11:537-61. [Crossref] [PubMed]
- Svoronos AA, Engelman DM, Slack FJ. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res 2016;76:3666-70. [Crossref] [PubMed]
- Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 2014;20:460-9. [Crossref] [PubMed]
- Guo L, Yang S, Zhao Y, et al. Global analysis of miRNA gene clusters and gene families reveals dynamic and coordinated expression. Biomed Res Int 2014;2014:782490 [PubMed]
- Guo L, Zhao Y, Zhang H, et al. Integrated evolutionary analysis of human miRNA gene clusters and families implicates evolutionary relationships. Gene 2014;534:24-32. [Crossref] [PubMed]
- Kamanu TK, Radovanovic A, Archer JA, et al. Exploration of miRNA families for hypotheses generation. Sci Rep 2013;3:2940. [Crossref] [PubMed]
- Griffiths-Jones S, Grocock RJ, van Dongen S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 2006;34:D140-4. [Crossref] [PubMed]
- Griffiths-Jones S, Saini HK, van Dongen S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res 2008;36:D154-8. [Crossref] [PubMed]
- Griffiths-Jones S. miRBase: microRNA sequences and annotation. Curr Protoc Bioinformatics 2010;Chapter 12:Unit 12.9.1-10.
- Hemmatzadeh M, Mohammadi H, Jadidi-Niaragh F, et al. The role of oncomirs in the pathogenesis and treatment of breast cancer. Biomed Pharmacother 2016;78:129-39. [Crossref] [PubMed]
- O'Bryan S, Dong S, Mathis JM, et al. The roles of oncogenic miRNAs and their therapeutic importance in breast cancer. Eur J Cancer 2017;72:1-11. [Crossref] [PubMed]
- Ortiz-Quintero B. Cell-free microRNAs in blood and other body fluids, as cancer biomarkers. Cell Prolif 2016;49:281-303. [Crossref] [PubMed]
- De Cecco L, Dugo M, Canevari S, et al. Measuring microRNA expression levels in oncology: from samples to data analysis. Crit Rev Oncog 2013;18:273-87. [Crossref] [PubMed]
- Callari M, Dugo M, Musella V, et al. Comparison of microarray platforms for measuring differential microRNA expression in paired normal/cancer colon tissues. PLoS One 2012;7:e45105 [Crossref] [PubMed]
- de Planell-Saguer M, Rodicio MC. Detection methods for microRNAs in clinic practice. Clin Biochem 2013;46:869-78. [Crossref] [PubMed]
- Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet 2012;13:358-69. [Crossref] [PubMed]
- Plummer PN, Freeman R, Taft RJ, et al. MicroRNAs regulate tumor angiogenesis modulated by endothelial progenitor cells. Cancer Res 2013;73:341-52. [Crossref] [PubMed]
- Patel Y, Lee J S, Chen H. Clinicopathological analysis of miRNA expression in breast cancer tissues by using miRNA In situ hybridization. J Vis Exp 2016;(112).
- Kappel A, Keller A. miRNA assays in the clinical laboratory: workflow, detection technologies and automation aspects. Clin Chem Lab Med 2017;55:636-47. [Crossref] [PubMed]
- Tian T, Wang J, Zhou X. A review: microRNA detection methods. Org Biomol Chem 2015;13:2226-38. [Crossref] [PubMed]
- Schwarzenbach H, da Silva AM, Calin G, et al. Data Normalization Strategies for MicroRNA Quantification. Clin Chem 2015;61:1333-42. [Crossref] [PubMed]
- Khan J, Lieberman JA, Lockwood CM. Variability in, variability out: best practice recommendations to standardize pre-analytical variables in the detection of circulating and tissue microRNAs. Clin Chem Lab Med 2017;55:608-21. [Crossref] [PubMed]
- Becker A, Thakur BK, Weiss JM, et al. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell 2016;30:836-48. [Crossref] [PubMed]
- Falcone G, Felsani A, D'Agnano I. Signaling by exosomal microRNAs in cancer. J Exp Clin Cancer Res 2015;34:32. [Crossref] [PubMed]
- Schwarzenbach H. Clinical relevance of circulating, cell-free and exosomal microRNAs in plasma and serum of breast cancer patients. Oncol Res Treat 2017;40:423-9. [Crossref] [PubMed]
- Madhavan D, Zucknick M, Wallwiener M, et al. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin Cancer Res 2012;18:5972-82. [Crossref] [PubMed]
- Sieuwerts AM, Mostert B, Bolt-de Vries J, et al. mRNA and microRNA expression profiles in circulating tumor cells and primary tumors of metastatic breast cancer patients. Clin Cancer Res 2011;17:3600-18. [Crossref] [PubMed]
- Fehm T, Braun S, Muller V, et al. A concept for the standardized detection of disseminated tumor cells in bone marrow from patients with primary breast cancer and its clinical implementation. Cancer 2006;107:885-92. [Crossref] [PubMed]
- Kallergi G, Politaki E, Alkahtani S, et al. Evaluation of Isolation Methods for Circulating Tumor Cells (CTCs). Cell Physiol Biochem 2016;40:411-9. [Crossref] [PubMed]
- Radom-Aizik S, Zaldivar F Jr, Leu S Y, et al. Effects of exercise on microRNA expression in young males peripheral blood mononuclear cells. Clin Transl Sci 2012;5:32-8. [Crossref] [PubMed]
- Atarod S, Smith H, Dickinson A, et al. MicroRNA levels quantified in whole blood varies from PBMCs. F1000Res 2014;3:183. [PubMed]
- Vaz C, Ahmad HM, Bharti R, et al. Analysis of the microRNA transcriptome and expression of different isomiRs in human peripheral blood mononuclear cells. BMC Res Notes 2013;6:390. [Crossref] [PubMed]
- Vaz C, Ahmad HM, Sharma P, et al. Analysis of microRNA transcriptome by deep sequencing of small RNA libraries of peripheral blood. BMC Genomics 2010;11:288. [Crossref] [PubMed]
- Su MW, Yu SL, Lin WC, et al. Smoking-related microRNAs and mRNAs in human peripheral blood mononuclear cells. Toxicol Appl Pharmacol 2016;305:169-75. [Crossref] [PubMed]
- Frampton AE, Fletcher CE, Gall TM, et al. Circulating peripheral blood mononuclear cells exhibit altered miRNA expression patterns in pancreatic cancer. Expert Rev Mol Diagn 2013;13:425-30. [Crossref] [PubMed]
- Zheng Y, Cui L, Sun W, et al. MicroRNA-21 is a new marker of circulating tumor cells in gastric cancer patients. Cancer Biomark 2011-2012;10:71-7. [Crossref] [PubMed]
- Yang B, Liu Z, Ning H, et al. MicroRNA-21 in peripheral blood mononuclear cells as a novel biomarker in the diagnosis and prognosis of prostate cancer. Cancer Biomark 2016;17:223-30. [Crossref] [PubMed]
- Chang CW, Wu HC, Terry MB, et al. microRNA expression in prospectively collected blood as a potential biomarker of breast cancer risk in the BCFR. Anticancer Res 2015;35:3969-77. [PubMed]
- Wang WS, Liu LX, Li GP, et al. Combined serum CA19-9 and miR-27a-3p in peripheral blood mononuclear cells to diagnose pancreatic cancer. Cancer Prev Res (Phila) 2013;6:331-8. [Crossref] [PubMed]
- Ma J, Lin Y, Zhan M, et al. Differential miRNA expressions in peripheral blood mononuclear cells for diagnosis of lung cancer. Lab Invest 2015;95:1197-206. [Crossref] [PubMed]
- Ma J, Guarnera MA, Zhou W, et al. A Prediction model based on biomarkers and clinical characteristics for detection of lung cancer in pulmonary nodules. Transl Oncol 2017;10:40-5. [Crossref] [PubMed]
- Markou A, Zavridou M, Sourvinou I, et al. Direct comparison of metastasis-related mirnas expression levels in circulating tumor cells, corresponding plasma, and primary tumors of breast cancer patients. Clin Chem 2016;62:1002-11. [Crossref] [PubMed]
- Leong SM, Tan KM, Chua HW, et al. Paper-based microRNA Expression profiling from plasma and circulating tumor cells. Clin Chem 2017;63:731-41. [Crossref] [PubMed]
- Jack RM, Grafton MM, Rodrigues D, et al. Ultra-specific isolation of circulating tumor cells enables rare-cell RNA Profiling. Adv Sci (Weinh) 2016;3:1600063 [Crossref] [PubMed]
- Ortega FG, Lorente JA, Garcia Puche JL, et al. miRNA in situ hybridization in circulating tumor cells--MishCTC. Sci Rep 2015;5:9207. [Crossref] [PubMed]
- Gasch C, Plummer PN, Jovanovic L, et al. Heterogeneity of miR-10b expression in circulating tumor cells. Sci Rep 2015;5:15980. [Crossref] [PubMed]
- Pixberg CF, Raba K, Muller F, et al. Analysis of DNA methylation in single circulating tumor cells. Oncogene 2017;36:3223-31. [Crossref] [PubMed]
- Brabletz S, Bajdak K, Meidhof S, et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J 2011;30:770-82. [Crossref] [PubMed]
- Krebs AM, Mitschke J, Lasierra Losada M, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol 2017;19:518-29. [Crossref] [PubMed]
- Zhang T, Wan JG, Liu JB, et al. MiR-200c inhibits metastasis of breast tumor via the downregulation of Foxf2. Genet Mol Res 2017;16. [PubMed]
- Banyard J, Chung I, Wilson AM, et al. Regulation of epithelial plasticity by miR-424 and miR-200 in a new prostate cancer metastasis model. Sci Rep 2013;3:3151. [Crossref] [PubMed]
- Chen P, Guo X, Zhang L, et al. MiR-200c is a cMyc-activated miRNA that promotes nasopharyngeal carcinoma by downregulating PTEN. Oncotarget 2017;8:5206-18. [PubMed]
- Chen J, Wang W, Zhang Y, et al. The roles of miR-200c in colon cancer and associated molecular mechanisms. Tumour Biol 2014;35:6475-83. [Crossref] [PubMed]
- Meng X, Muller V, Milde-Langosch K, et al. Circulating Cell-Free miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Adv Exp Med Biol 2016;924:3-8. [Crossref] [PubMed]
- Shao Y, Geng Y, Gu W, et al. Prognostic role of tissue and circulating microRNA-200c in malignant tumors: a systematic review and meta-analysis. Cell Physiol Biochem 2015;35:1188-200. [Crossref] [PubMed]
- Le MT, Hamar P, Guo C, et al. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest 2014;124:5109-28. [Crossref] [PubMed]
- Madhavan D, Peng C, Wallwiener M, et al. Circulating miRNAs with prognostic value in metastatic breast cancer and for early detection of metastasis. Carcinogenesis 2016;37:461-70. [Crossref] [PubMed]
- Lowery AJ, Miller N, Dwyer RM, et al. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer 2010;10:502. [Crossref] [PubMed]
- Xiong DD, Lv J, Wei KL, et al. A nine-miRNA signature as a potential diagnostic marker for breast carcinoma: An integrated study of 1,110 cases. Oncol Rep 2017;37:3297-304. [Crossref] [PubMed]
- Cheng Y, Xiang G, Meng Y, et al. MiRNA-183-5p promotes cell proliferation and inhibits apoptosis in human breast cancer by targeting the PDCD4. Reprod Biol 2016;16:225-33. [Crossref] [PubMed]
- Song C, Zhang L, Wang J, et al. High expression of microRNA-183/182/96 cluster as a prognostic biomarker for breast cancer. Sci Rep 2016;6:24502. [Crossref] [PubMed]
- Shimono Y, Mukohyama J, Nakamura S, et al. MicroRNA regulation of human breast cancer stem cells. J Clin Med 2015;5:2. [Crossref] [PubMed]
- Peng Q, Zhang X, Min M, et al. The clinical role of microRNA-21 as a promising biomarker in the diagnosis and prognosis of colorectal cancer: a systematic review and meta-analysis. Oncotarget 2017;8:44893-909. [PubMed]
- Hernandez YG, Lucas AL. MicroRNA in pancreatic ductal adenocarcinoma and its precursor lesions. World J Gastrointest Oncol 2016;8:18-29. [Crossref] [PubMed]
- Lu J, Xie F, Geng L, et al. Potential Role of MicroRNA-210 as biomarker in human cancers detection: a meta-analysis. Biomed Res Int 2015;2015:303987 [PubMed]
- Bahrami A, Aledavood A, Anvari K, et al. The prognostic and therapeutic application of microRNAs in breast cancer: Tissue and circulating microRNAs. J Cell Physiol 2018;233:774-86. [Crossref] [PubMed]
- Moles R. MicroRNAs-based therapy: a novel and promising strategy for cancer treatment. Microrna 2017;6:102-9. [Crossref] [PubMed]
- Riethdorf S, Fritsche H, Muller V, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res 2007;13:920-8. [Crossref] [PubMed]
- Yoo B, Kavishwar A, Wang P, et al. Therapy targeted to the metastatic niche is effective in a model of stage IV breast cancer. Sci Rep 2017;7:45060. [Crossref] [PubMed]
- Kim J, Siverly AN, Chen D, et al. Ablation of miR-10b suppresses oncogene-induced mammary tumorigenesis and metastasis and reactivates tumor-suppressive pathways. Cancer Res 2016;76:6424-35. [Crossref] [PubMed]
- Wang N, Chen P, Huang LP, et al. Prognostic significance of microRNA-10b overexpression in breast cancer: a meta-analysis. Genet Mol Res 2016;15:gmr.15027350.
- Gulyaeva LF, Kushlinskiy NE. Regulatory mechanisms of microRNA expression. J Transl Med 2016;14:143. [Crossref] [PubMed]


