
Circulating DNA-based lung cancer diagnostics and follow-up: looking for epigenetic markers
Introduction
Lung cancer (LC) remains the world’s leading cause of cancer-associated mortality (1). Due to the late manifestation of symptoms and low sensitivity of chest radiography used as screening technique most LCs (about 85%) are diagnosed in patients with either locally advanced disease and/or developed distant metastases, which results in a tremendously low 5-year survival rate (4% in case of a metastatic disease) (2). By contrast, 5-year survival rate of early stage LC patients exceeds 50%. Recently, low dose computed tomography (LDCT) was shown to be a more promising screening tool, but its implementation into the routine practice is hurdled by high false positive rate (fewer than 5% of discovered nodules are malignant), high cost and unexplored health risks (3). False positive CT results entail follow up procedures that are invasive, costly, and have associated morbidity and mortality. Likewise, magnetic resonance imaging (MRI) is a costly method with high false positive rates and is mostly applied to confirm existing diagnoses. Histological analysis of samples collected using bronchoscopy biopsies and fine needle aspirations provides valuable diagnostic information but has drawbacks due to procedure invasiveness.
Alongside early diagnostics another major battlefield in the war against LC is the assignment of appropriate and effective treatment regimes. LC is a diverse group of histologically and clinically distinct malignancies and therefore the effect of therapeutic measures differs dramatically on a case-to-case basis even for superficially similar tumors (4). Up to 30% of surgically treated stage I patients are lost to recurrent disease (5). Identification and clinical application of multiple noninvasive or tissue-based molecular biomarkers can assist in understanding the taxonomy and molecular landscape of LC necessary to identify (stage I) patients at high-risk for recurrence who may benefit from adjuvant chemotherapy or innovative immunotherapy regimens (5). Similarly, immunotherapies targeted at specific genetic alterations such as EGFR mutations and ALK fusions are known to improve outcomes for a subset of patients even with advanced stage of LC (6,7). Recent promising results demonstrate that genomic landscape of lung tumours shapes response to antibody targeting programmed cell death-1 (anti-PD-1) therapy, suggesting that exome-guided neoantigen identification may improve treatment responses (8).
Thus, new strategies for preclinical screening and monitoring of post therapy relapses based on selected “omics” biomarkers may be instrumental in relieving the burden of LC.
Cell-free circulating DNA (cfDNA) biomarkers
Complementary diagnostics using blood-derived molecular DNA markers is one attractive solution to the problem. cfDNA isolated from the blood of cancer patients contains DNA fragments shed from tumor cells and provides a convenient and minimally invasive access to the molecular portrait of cancer (9,10). Recently, cell-surface-bound fraction of circulating DNA (csbDNA) was proposed as an additional source of material (11-13). Cell-free circulating tumour DNA (ctDNA) in the blood plasma/serum presents a surrogate for the entire cancer genome (including the clonal landscape of the primary tumour and metastases) which gives it a unique appeal as liquid biopsy that provides “real-time” information about the evolution of tumor genome essential for planning the precision treatment regime. Our knowledge of cfDNA origins, mechanism and rate of release is still incomplete. The main sources are proposed to be the necrotic and apoptotic cell death (either before or post macrophage engulfment and digestion) along with active secretion by living cells (9,14,15). In any case, all tissues of the multicellular body have the potential to contribute to the collective information of the circulating genome. Another reason to consider ctDNA for liquid biopsy is concerned with its rapid clearance from bloodstream with half-life of approximately one hour followed by a slow phase with half-life of 13 hours as shown by the study of fetal cfDNA kinetics (16). These properties of cfDNA make it a useful tool for detection of residual cancer disease and recurrence during the post-treatment follow-up.
KRAS oncogene mutation was the first cfDNA marker detected in blood of cancer patients (17). Diagnostic tests for EGFR and KRAS mutations in cfDNA are now commercially available and can be used as prognostic indicators in advanced NSCLC patients. Meta-analysis of recent data demonstrated that EGFR mutations in ctDNA predicted better progression-free survival in advanced NSCLC patients treated by EGFR-TKIs (18). The feasibility of using tumour-specific mutations in ctDNA in diagnosis of cancer and monitoring of response to therapy has been widely demonstrated, although, the high individual variability of mutational landscape of tumour DNA makes it a labor-intensive approach (19).
Advantages of ctDNA methylated markers
Methylation of cytosines on position 5 in CpG dinucleotides is one of the most important epigenetic modifications in DNA of eukaryotic cells necessary for cellular differentiation and normal development of tissues and organs. Changes in DNA methylation are a common feature of most cancer types and occur early and consistently in cancer development, thus making aberrant DNA methylation a broadly applicable marker of ctDNA in blood alongside mutations, deletions, loss of heterozygosity, etc. (20). However, the consistency of methylation events in cancer suggests them a somewhat more reliable option. Age-, gender- and smoking-associated changes in methylation profile are also known to occur and should be considered when methylation alterations are used for diagnostic purposes (21). Implications of these findings for the study of ctDNA methylation are currently being reviewed.
Previously, aberrant DNA methylation has been strongly implicated in cancer initiation and progression (21,22). Hypermethylation of CpG-rich regions in gene promoters called CpG islands (CGIs) may induce repression of individual tumour suppressor genes, while global hypomethylation contributes to tumourigenesis through the promotion of genomic instability and activation of oncogenes. In non-small cell lung cancer (NSCLC) hypermethylation of tumour suppressor genes can serve as an indication of stage and histological type, aiding in assessing survival prognosis, disease progression and recurrence (23). Most of the studies to date have been conducted on cfDNA to validate previous discoveries of CpG island hypermethylation of certain single-copy genes (24-26). An alternative strategy is to look at hypomethylation of genetic mobile elements, which are scattered around the human genome as multiple repeated copies and normally silenced by methylation (27,28). Detection of repetitive DNA should have improved sensitivity compared with a single-copy gene assay. Hypomethylation of LINE-1 retrotransposons in lung tumours (29-31) and blood cells of LC patients (32,33) has been described as one of the key hallmarks of carcinogenesis. Moreover, the degree of LINE-1 hypomethylation was associated with clinical features of the disease and survival prognosis (34,35). Therefore, hypomethylation of repetitive elements could be used as a screening, diagnostic and prognosic biomarker in LC.
As mentioned before, cfDNA represents a mixture of molecules originating from different tissues, with ctDNA accounting for as low as 0.05% of total cfDNA or less in many cancer patients, especially at the early stages of the disease (36). High accuracy of discrimination between unmethylated and methylated alleles is possible due to the development of the bisulfite conversion technique based on chemical treatment of DNA strands by sodium bisulphite that converts unmethylated cytosines to uracils. Methylation-specific PCR (MSP) of bisulphite converted DNA allows for a highly specific detection of a single methylated allele over a background of more than 1,000-fold excess of unmethylated alleles, which is crucially important for the study of ctDNA.
These properties make aberrant methylation of ctDNA a promising cancer biomarker, and recent high throughput investigations have demonstrated the correspondence between the changes in methylation profiles of ctDNA and DNA from paired tumor tissue (37-40). However, DNA methylation-based biomarkers have not been incorporated into commercially available assays for in vitro diagnostics until very recently, and currently Epi ProColon (Epigenomics AG, Berlin, Germany) is the only FDA approved ctDNA assay for colorectal cancer screening based on qPCR detection of hypermethylated Septin9 in the ctDNA derived from blood plasma. A prospective LC screening assay from the same company named Epi ProLung BL is based on qPCR detection of hypermethylated SHOX2 and PTGER4 in DNA from bronchoalveolar fluid obtained from patients during bronchoscopy. Notably, the authors have also recently demonstrated the potential utility of methylated SHOX2 and PTGER4 in plasma cfDNA for LC screening (41) (Table 1).
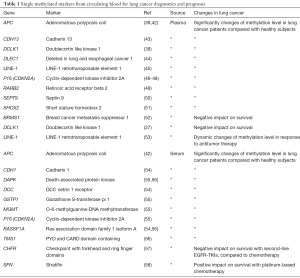
Full table
Technical aspects of the methylated cfDNA analysis
The long and winding road of cfDNA-based methylated liquid biopsy markers into clinical practice can be attributed to the challenges inherent to the nature of cfDNA. It is present in the circulation in a low quantity of less than 10 ng per mL of blood plasma, and low quality, inasmuch as it is highly fragmented and mostly consists of roughly 180 bp molecules. So far, several reviews aimed to determine the optimal pre-analytical considerations necessary to optimize cfDNA yield and quality, establish protocols for cfDNA analysis and suggest guidelines for translation of cfDNA analysis into routine clinical practice (9,59-61). These reports defined various parameters for optimal blood sample handling before cfDNA isolation based on literature data and proposed confirmatory experiments as a first step in this direction. In brief, when developing a methylated ctDNA marker assay one should first standardize sample preparation: a choice must be made between plasma and serum, storage temperature and time interval between blood sampling and sample processing should be optimized, suitable DNA extraction and purification methods should be selected. In order to minimize the loss of cfDNA fragments during isolation, a broad range of commercial kits designed to recover low molecular weight nucleic acids is available, although when compared they demonstrate vastly different efficiency and reproducibility (62-64). Hulbert et al developed the methylation-on-beads (MOB) technique, which reduces sample loss thereby potentially increasing sensitivity (65).
Cytosine methylation detection techniques based on bisulfite conversion
Second step in setting up a cfDNA assay is to select an effective method for discriminating between methylated and unmethylated CpG sites. The treatment of cfDNA with sodium bisulfite has become the most widely used method, easily combined with various downstream detection technologies, including PCR-based assays, pyrosequencing, dideoxy-sequencing, high resolution melting, single-strand conformation polymorphism analysis, microarrays, next-generation sequencing (NGS), MALDI-TOF mass spectrometry, etc. Quantitative MSP assays and digital PCR-based approaches have high analytical sensitivity of detection of rare methylated alleles in the presence of excess unmethylated cfDNA, making them suitable for the analysis of ctDNA. MSP initially only allowed for a qualitative estimation of methylation of the region of interest. Quantitative MSP using a Taqman probe (MethyLight) enabled the detection of methylated allele in a 10,000-fold excess of unmethylated alleles thus providing a 10-fold increase of sensitivity compared to conventional MSP. A further evolution of this approach is digital MethyLight (dMethyLight) which is based on compartmentalization of DNA templates over multiple reaction wells, allowing for detection of single methylated alleles and quantitative analysis via counting of positive wells (66). Droplet digital MethyLight (ddMethyLight) further improves on the sensitivity of MethyLight assay by using droplets instead of wells used in dMethyLight, increasing effective dilution factor by an order of magnitude (67). In the recent years other highly sensitive technologies have been developed, such as Methyl-BEAMing combining emulsion dPCR with magnetic beads and flow cytometry to achieve sensitivity equivalent to dPCR (68), or RainDrop digital PCR (69). An approach called DREAMing (Discrimination of Rare EpiAlleles by Melting) was proposed as a solution to the detection and assessment of epigenetic heterogeneity of ultra-rare epiallelic variants present in liquid biopsies (70). DREAMing relies on semi-limiting dilution of DNA samples and precision melt curve analysis to quantify the methylation density of the heterogeneously-methylated templates.
A fundamentally different approach to the analysis of methylated CpGs is pyrosequencing—sequencing-by-synthesis system that relies on the luminometric detection of pyrophosphates released as nucleotides are incorporated into the extended strand. This technique provides quantitative data of the methylation status of multiple individual CpGs within a region of interest in bisulfite-treated DNA (71). Usefulness of pyrosequencing-based methods on ctDNA samples is limited compared with PCR-based assays by the gradual decay of the accuracy of evaluation of methylation status for CpG sites based on the distance from the 3' end of the primer.
Nevertheless, there are several significant drawbacks associated with the chemical conversion of cytosines. One of the main drawbacks of bisulfite conversion is the introduction of breaks in the DNA, leading to a dramatic loss of already highly fragmented cfDNA. Bisulfite treatment reduces sequence complexity, puts constrains on primer design for PCR amplification, and disallows the distinction between 5-methylcytosines and 5-hydroxymethylcitosines (72), leading to false-positives in downstream analyses. Finally, incomplete conversion of either methylated or unmethylated cytosines leads to false-positive and false-negative results even when using commercially available kits (73). Therefore, a number of bisulfite-independent methods have been developed aiming to improve the locus-specific CpG methylation detection.
Other methods and their combinations
Affinity enrichment-based methods utilize specific antibodies recognizing methylated cytosines or methyl-binding proteins to selectively recover methylated DNA. Examples include MethylCpG Binding Domain MBD2 proteins (MBD, also termed Methyl Cap), methylated DNA immunoprecipitation (MeDIP) and methylated CpG island recovery assay (MIRA) (74-76). MBD fusion proteins bind specifically to dsDNA that is methylated at CpG sites on both strands. They demonstrate a bias for high CpG densities and preferentially recover methylated CGIs (60). Commercially available MBD affinity kits use MBD proteins fused with glutathione-S-transferase (GST) (MethylMagnet, RiboMed; MethylQuest, Millipore; Methyl-Cap, Diagenode) or with His6 (hexahistidine-tag or polyhistidine-tag) affinity tags immobilized to magnetic beads (MethylMiner, Life Technologies; MethylCollector, Active Motif). These techniques can be combined with quantitative PCR to perform locus-specific assessment of methylation or with sequencing to obtain whole-genome methylation profiles. Low recovery of methylated DNA is the main disadvantage of this approach, alongside its inability to target single CpG sites.
Another group of methods employs methylation-sensitive restriction enzymes that cleave at sites with unmethylated CpGs, therefore in the following PCR only the methylated alleles are amplified. Methylation-sensitive restriction enzymes can be used in combination with different detection techniques, such as combined bisulfite restriction analysis (COBRA), differential methylation hybridization (DMH), HpaII tiny fragment enrichment by ligation-mediated PCR (HELP), and methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) (77). In order to circumvent the major disadvantage of this approach, stemming from the ability of the enzyme to only recognize a set pattern of CpG sites, another approach called Methylation Specific Nuclease-assisted Minor-allele Enrichment (MS-NaME) was recently developed (78). This technique utilizes oligonucleotide probes that direct double-strand-specific DNA nuclease (DSN) to multiple targets in bisulfite-treated DNA followed by amplification of the targeted regions, resulting in the enrichment of multiple methylated or unmethylated rare epigenetic alleles. Additional examples of the integrative approach are the nucleosome occupancy and methylome sequencing (NOMe-seq) method which allows to simultaneously determine the nucleosome occupancy and DNA methylation. In NOMe-seq, nuclei are treated with GpC Methyltransferase (M.CviPI) before bisulfite conversion and sequencing.
To sum up, a plethora of methods and their modifications utilised for the detection of DNA methylation provides an array of potential solutions for biomarker research, addressing the needs of a wide variety of studies in LC, but simultaneously rendering the comparison of results very difficult, and creating a major obstacle for conducting meta-analyses (79).
Discovery and validation of epigenetic markers
Third critical step in the development of an effective methylated ctDNA-based assay is to select the valid markers (61). Two approaches for marker selection have been utilized so far. One approach entails the selection of tumour tissue-associated candidate genes, followed by development and validation of sensitive and specific assay using representative groups of patients and controls. Another approach relies on genome-wide methylation profiling techniques, such as array hybridization and NGS of cfDNA. The former method has instigated the evaluation of a number of potentially useful epigenetic signatures as markers applicable for LC diagnosis, staging, prognosis and assessment of the response to therapy. Some of the candidates are currently undergoing validation studies. Notably, this approach was used in the development of the successful Epi ProColon assay. In order to select the proper strategy for the development of novel biomarkers it is imperative to understand if the benefits, potential biases and resource requirements of a particular technique are suitable to meet the research objectives of the study (61,80,81).
Genome-wide methylation profile analysis
Studies based on genome-wide methylation profiling of cfDNA remain scarce due to technical challenges such as low starting amounts of cfDNA, further aggravated by the loss of material during sample processing, DNA extraction, bisulfite conversion and library preparation.
Assessment of DNA methylation on a genome level was initially conducted by a combination of microarray hybridization with either digestion of DNA by methylation sensitive enzymes, affinity purification or bisulfite conversion (82). One of the most widely used array-based approaches for methylation profiling is the Infinium HumanMethylation450 BeadChip Array (Illumina) or 450k, which is able to detect the methylation status of 485,000 individual CpGs from reference genes and CGIs (83). The reason behind the popularity of the 450k platform are the low cost of batch processing of the samples and ease of data analysis using publicly available R packages. Array-based methodologies are ideal for first-pass methylation profiling; however, the requirement for a large amount of input DNA (hundreds of ng) precludes their use for widespread assessment of DNA methylation in liquid biopsies. Still, encouraging results from a recent pilot study showed that genome-wide profiles of DNA methylation in ctDNA are consistent with corresponding tumor tissues (84).
Whole genome bisulfite sequencing (WGBS) is perhaps the most powerful method to explore the methylome with the potential to identify the methylation status of every CpG site in the genome. WGBS libraries are prepared similarly to regular whole genome sequencing, with the additional step of bisulfite conversion (85). The critical determinants for applying high-throughput WGBS to epigenome-wide association studies for cfDNA-based cancer marker discovery are reviewed elsewhere (81). Targeted sequencing using panels consisting of tens to hundreds of select genes is a more appealing and cost-effective solution for ctDNA analysis. Recently whole-genome bisulfite sequencing from nanogram quantities of input cfDNA has been enabled by novel methods such as T-WGBS (transposase-based library construction) and PBAT (post-bisulfite adaptor tagging, which can be performed with as little as 125 pg of input DNA) (86-89). Study of thousands of hypermethylated CGIs in cfDNA using methylated CpG tandems amplification and sequencing (MCTA-Seq) was performed by Wen et al. in tissue and plasma samples from hepatocellular carcinoma (HCC) patients and healthy controls, resulting in the identification of a panel of four cancer-specific genes (RGS10, STSIA6, RUNX2 and VIM) (Table 2) (89).
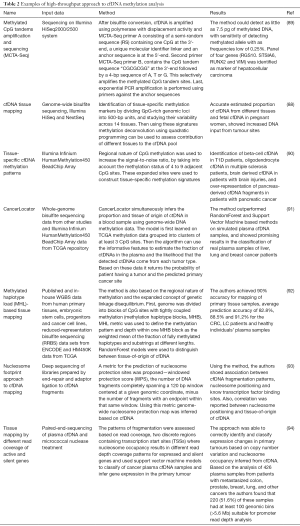
Full table
Distinct advantage of genome-wide DNA methylation assays is the option to apply innovative bioinformatic solutions to data analysis, as evidenced by a great number of recent investigations reporting newly developed approaches for evaluating the proportion and the tissue-of-origin of tumor-derived cell-free DNA in blood samples with the general aim of developing a universal cancer detection technique (Table 2). The underlying principle of cfDNA tissue mapping is based on the genome-wide bisulfite sequencing of plasma cfDNA and methylation deconvolution of the sequencing data (88). Stretches of adjacent CpG sites are used as tissue-specific methylation markers to increase signal-to-noise ratio and improve the sensitivity of the assay. Sun et al. [2015] (88) estimated the proportion of cfDNA contributed by different tissues and showed that an abnormally high presence of cfDNA from a specific tissue may indicate it as the tumor site. Lehmann-Werman et al. [2016] (90) applied the same rationale to the diagnosis of pancreatic cancer (90). With the help of the Illumina 450k methylation arrays (HM450K) platform they showed that roughly 50% of pancreatic cancer patients demonstrated a substantial over-representation of pancreas-derived cfDNA fragments compared with healthy subjects. Recent study from Kang et al. proposed a probabilistic method, CancerLocator, which exploits the diagnostic potential of cfDNA to determine not just the presence but also the location of the tumors, based on methylation signatures associated with tissue-specific cancers (91). Guo et al. [2017] performed an exhaustive search of tissue-specific methylation haplotype blocks (MHBs) across the entirety of the genome and proposed a methylated haplotype load (MHL) block-level metric for systematic discovery of informative markers. By using an original analytical framework and identified markers, the authors demonstrated the ability to accurately identify the tissue origin of cfDNA and classify plasma samples from LC and colorectal cancer patients (92).
Another innovative approach to cfDNA tissue mapping comes from the discovery of the direct association between cfDNA fragmentation patterns and nucleosome positioning (93). Ulz et al. [2016] performed whole-genome sequencing of plasma cfDNA and identified two discrete regions containing transcription start sites (TSSs) where nucleosome occupancy resulted in different read depth coverage patterns for expressed and silent genes (94). By employing machine learning the authors were able to classify expressed cancer driver genes in regions with somatic copy number gains with high accuracy.
Early LC screening
So far, a significant number of candidate tumour tissue-associated genes were selected and thoroughly validated, yielding the biomarkers listed in Table 1 and reviewed elsewhere (95,96). The list includes hypermethylated SHOX2 (38,39,41,51), RASSF1A (4,56,97-99), RARB2 (49,97), P16 (46-48,55,98), MGMT (54,55,100), death-associated protein kinase (DAPK) (55,100,101), APC (39,42,54) and DLEC1 (39,44). Notably, detection of methylated markers in blood plasma of smokers with COPD, which have a high risk of developing LC, predicted LC 3–18 months before its clinical diagnosis (99). Two genes, P16 and ESR1, are methylated in the peripheral blood DNA of stage I LC patients, providing higher sensitivity of LC detection in the same patients than CEA (102).
Diagnostic efficiency of individual gene assays is generally lower compared with a panel of two or more methylated markers previously reported to have high diagnostic potential (Table 3). Their potential for cancer screening is at its highest if used as complementary tests to improve the diagnostic accuracy of LC risk assessment by low-dose computed tomography (LDCT) screening (Figure 1). The National Lung Screening Trial (USA) demonstrated a 20% reduction in LC mortality using LDCT screening. If confirmed in a validation study, the developed panels of methylation markers could be used as a companion test for LDCT screening, identifying patients at high risk for LC, reducing false positive results, preventing unnecessary tests, and improving the detection and diagnosis of LC at the early stages. Recently a case-control study of subjects with suspicious nodules revealed by LDCT imaging was conducted using quantitative methylation-specific real-time PCR and Methylation on Beads for six genes (SOX17, TAC1, HOXA7, CDO1, HOXA9, and ZFP42) (105), selected for the study using The Cancer Genome Atlas (TCGA) as having high prevalence of DNA methylation in lung squamous and adenocarcinoma, but not in normal lung tissue. A combination of three best individual genes has sensitivity and specificity of 98% and 71% using sputum and 93% and 62% using plasma. Independent blinded random forest prediction models combining gene methylation with clinical information correctly predicted LC in 91% of subjects using TAC1, HOXA17 and SOX17 in sputum and 85% of subjects using CDO1, TAC1 and SOX17 in plasma. Another signature of four effective markers (HOXD10, PAX9, PTPRN2, and STAG3) was reported by Wielscher et al. [2015] (106) who used a successive selection by genome-wide methylation screening, methylation-sensitive restriction enzyme digestion and enrichment of methylated DNA and parallel qPCR for the panel development. The four-marker model yielded an AUC of 0.85 with sensitivity of 97% and a specificity of 73%.
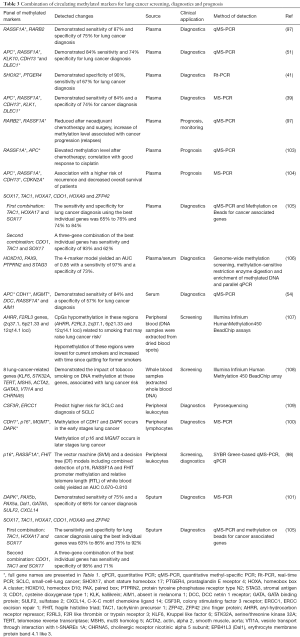
Full table
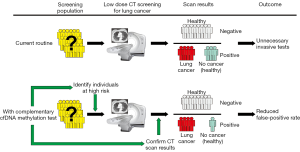
Methylated SHOX2 has been proposed as a putative ctDNA marker for LC diagnosis in a number of studies with median sensitivity (60%) and high specificity (90%) (51). Three independent case-control studies examined a total of 330 plasma cfDNA specimens for methylated (SHOX2), prostaglandin E receptor 4 gene (PTGER4), and forkhead box L2 gene (FOXL2) and a panel of SHOX2 and PTGER4 and yielded promising results (41). Importantly, a validation study with 172 patient samples demonstrated high performance in distinguishing patients with LC from subjects without malignancy. At a fixed specificity of 90%, sensitivity for LC was 67%; at a fixed sensitivity of 90%, specificity was 73%. At maximum sensitivity, this assay shows a reasonably high specificity that can provide substantial reduction in the false-positive rate of LDCT. If adopted with high specificity, the test may prove be useful as a screening test directing patients to LDCT screening.
Compared to hypermethylation of tumour suppressor genes, hypomethylation of repetitive elements as Alu, LINE-1 (31,33) in cfDNA have attracted less attention and has not been studied thoroughly. Many studies have reported LINE-1 hypomethylation in various cancer tissues, including LC tumours (29). Global genome hypomethylation in LC tissues in known to contribute to genomic instability (31), and was associated with poor outcome (79) and overexpression of aberrant transcripts such as ΔNp73 (79). Based on the data presented by Hoshimoto et al., increase of LINE-1 hypomethylation was observed in the cfDNA from serum of stage III or IV malignant melanoma patients, compared to healthy donors (110). Recent study evaluated the potential of LINE-1 hypomethylation in plasma cfDNA as a blood biomarker for early stage CRC detection (111). Due to the low amount of tumour DNA in the circulation, large volume of plasma is needed to ensure the sensitivity of the ctDNA-based assay; for instance, mSEPT9-based test Epi proColon requires 3.5 mL of plasma. Conversely, with the abundance of LINE-1 copies in the human genome, only 0.5 mL of plasma enables to quantify its absolute methylation level (111). Our group performed MIRA coupled with qPCR-based quantitation to assess the integral methylation level of LINE-1 promoters in the csbDNA of NSCLC patients and healthy controls (Table 1). Deep sequencing of amplicons revealed that hypomethylation of LINE-1 promoters in csbDNA of LC patients is more pronounced for the human-specific L1Hs family. Statistical analysis demonstrated significant difference in L1Hs promoter methylation between cancer patients and healthy individuals (AUC =0.69).
Reviewing these and earlier studies, we should stress the urgency for the validation of best performing methylation markers selected from different studies in multi-centric international collaborations with larger cohorts, potentially resulting in an optimized gene panel. Adequate representation of histologic subgroups, TNM subclasses for LC, and inclusion of non-malignant and pre-cancerous states, such as chronic obstructive pulmonary disease (COPD), fibrotic interstitial lung diseases (ILDs) and other non-cancer patient groups, are necessary for the success of the validation study. Evaluation of selected markers using universal detection protocols in a single validation study should allow assess their true efficiency and adequately compare the potential of different markers, resulting in the development of valid biomarker panels (79,112).
Monitoring toxicity of chemotherapy and tumour resection efficiency
Early detection of resistance to chemotherapy treatment is imperative for improving patient outcomes, by minimizing the unnecessary toxicity and accelerating the transition to alternative therapy regimens. ctDNA is a promising new approach for monitoring of changes in tumor burden in response to therapy. Elevated ctDNA levels were shown to precede clinical establishment of progressive disease (113), thus ctDNA may provide an early marker of cancers resistance to therapy.
The potential of SHOX2 in monitoring treatment effectiveness in LC patients receiving chemotherapy was evaluated (114). Patients responding to therapy (17/36) showed a decrease of methylated SHOX2 in plasma at the first post-treatment blood draw 7–10 days after administration of chemotherapy, while for non-responders (8/19) the decrease was significantly smaller. These data suggest that methylated plasma SHOX2 is able to identify patients who will benefit from chemotherapy early and with a high accuracy. The methylation level of two other tumour suppressor genes—RASSF1A and RARB2—was earlier shown to be associated with LC development by different groups. RASSF1A and RARB2 methylation indices in cfDNA and csbDNA were found to be indicative of treatment response in non-small-cell LC patients (97) (Table 3). Both RASSF1A and RARB2 showed a decrease in methylation index after 2 courses of neoadjuvant chemotherapy, and then a further decrease following surgery. During the 9 months of after-treatment follow-up, 5 out of 26 patients had a relapse, all of them having the plasma methylation of at least one gene return to before treatment levels, while no patients without recurrence showed an increase in the methylation index (97) (Table 3). Decreased tumor volume was associated with the decrease in methylated ctDNA markers when measured 10–15 days post-therapy which evidenced of tumour chemosensitivity. In contrast, when methylation of APC1 and RASSF1A measured in ctDNA of LC patients immediately prior to the administration of chemotherapy and again 24 h after, when DNA released from dying cells peaks, was compared, an increase in circulating methylated RASSF1A/APC1 was shown to be associated with chemosensitivity and complete or partial response to treatment (103). In a very recent study authors demonstrated that quantitative analysis of total plasma cfDNA and plasma APC/RASSF1A methylation collectively provided a real-time synchronous rapid monitoring indicator of therapeutic outcomes of advanced LC. Four parameters were assessed: methylation level before chemotherapy (meth0 h), methylation level 24 h after chemotherapy (meth24 h), total plasma DNA concentration before chemotherapy (DNA0 h), and total plasma DNA concentration 24 h after chemotherapy (DNA24 h) (103). Meth24 h > meth0 h for at least one gene and DNA24 h/DNA0 h ≤2 were defined as criteria for better tumor response and fewer adverse events with a high correct prediction rate (84.7%). These fluctuations underscore the importance of carefully characterizing ctDNA kinetics in response to chemotherapy as a part of bringing the biomarkers to the clinic for patient monitoring.
The persistence of cancer DNA in blood after tumor has been removed likely reflects residual tumor tissue in the body and indicates poor prognosis (36). Tumour-specific methylation is less variable across tumors than mutation therefore their quantification in blood plasma provides a less labour-intensive approach that is more appealing for clinical use. The presence of tumor-specific methylated sequences that have been shown to decrease in the blood of LC patients following surgery is compiled in Table 3.
Methylation markers of cancer prognosis
Gene methylation patterns in tumor tissue can be indicative of tumor aggressiveness and the likelihood of recurrence after surgical resection and/or chemotherapy due to residual disease (115). Numerous studies have linked methylation of individual genes (116-118) and gene panels (119-121) in cancer tissue with patient survival. Recent study from the group of Prof. Zhang evaluated the utility of DNA methylation signatures for differentiating between tumor and normal tissue for four common cancers (breast, colon, liver, and lung) using machine learning on whole-genome methylation data from TCGA (122). The study demonstrated the potential of using methylation signatures to identify cancer tissue of origin and predict prognosis.
ctDNA has a short half-life (~2 h), and its persistence in the blood following surgery has been linked to poor prognosis (36). Prognostic biomarkers are urgently needed to distinguish patients who are cured with surgery alone, from those at high risk of disease recurrence who may benefit from adjuvant chemotherapy (61). The prognostic significance of gene promoter ctDNA methylation has been described in several studies. Li et al. (118) performed a meta-analysis of cohort studies to determine whether promoter methylation of the DAPK gene contributes to the pathogenesis of NSCLC. Subgroup analysis based on sample source discovered that DAPK gene methylation was implicated in the pathogenesis of NSCLC in both blood and tissue subgroups (all P < 0.05). Detection of methylated breast cancer metastasis suppressor-1 (BRMS1) and (sex determining region Y)-box 17 (SOX17) in operable and advanced NSCLC, was shown to have a negative impact on survival (37,52). DCLK1 methylation was also associated with shorter survival (27). In contrast, SFN (14-3-3 Sigma) promoter methylation correlated with a reduced risk of death (58) (Table 1).
Conclusions
So far, a number of studies have supported the potential of ctDNAs methylation for the development of the minimally invasive tests for cancer diagnostics. Changes in the methylation of ctDNA in the blood plasma and serum can be used as sensitive and specific markers for LC diagnostics, prognosis and monitoring of the response to therapy. However, further detailed studies are required to make potential biomarkers a routine tool in the laboratory medicine/clinical practice. Pre-analytical methods and analysis of ctDNA methylation profiles should be standardized, automated and certified in order to enable rapid and reliable detection and quantification. As the sensitivity of analytical techniques increases, detection of low levels of methylated DNA becomes possible, giving us the opportunity to detect early stages of cancer or minimal residual disease. Therefore, LC-specific methylation has the potential to be used as a minimally invasive biomarker in combination with LDCT to direct individuals to LDCT screening and minimize false positive diagnoses. The use of high-throughput and robust methodologies in diagnostic laboratories should advance the selection of the informative biomarkers. Recently, a multicenter benchmark study was carried out by 19 research groups from seven countries, assessing the currently available promising DNA methylation assays for biomarker validation studies (123). The results of the study endorse locus-specific DNA methylation assays as mature technology ready for widespread use in biomarker development and clinical applications. Another recent study declared uniquely mappable WGBS data to be the most reproducible and accurate measurement of global DNA methylation levels (124). WGBS was previously considered too costly for epidemiology studies with large cohorts of samples however, using multiplexing by indexed barcodes the costs of WGBS can be lowered significantly to improve the accuracy of global DNA methylation assessment for human studies. Most candidate ctDNA methylation markers are discovered in small retrospective cohorts or case-control studies, and few are validated in independent studies. To achieve the required degree of optimization, ctDNA methylation marker, panels and/or signatures should be developed and validated in large prospective cohort studies and screening populations. Clinical trials of ctDNA methylation as an early indicator of LC, prognostic marker or target for surrogate DNA demethylating activity are currently ongoing (61). Additional research is needed to explore the pathologic significance of the cfDNA, emerging from tumor cells into the circulation.
Acknowledgments
Funding: The study was supported by the fundamental research program of the Presidium of the Russian Academy of Sciences “Fundamental research for the development of biomedical technologies” (No. 2014-208), the program of the Presidium of the Russian Academy of Sciences “Molecular and Cellular Biology” and the Russian Foundation for Basic Research (No. 17-29-06002).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Heidi Schwarzenbach) for the series “Technologies in Liquid Biopsies - Potential applications in Medicine” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.02.08). The series “Technologies in Liquid Biopsies - Potential applications in Medicine” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- National Cancer Institute Data, accessed on May, 19, 2015. Available online: http://seer.cancer.gov/statfacts/html/lungb.html
- Thomas A, Liu SV, Subramaniam DS, et al. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat Rev Clin Oncol 2015;12:511-26. [Crossref] [PubMed]
- Prosch H. Implementation of lung cancer screening: promises and hurdles. Transl Lung Cancer Res 2014;3:286-90. [PubMed]
- Robles AI, Harris CC. Integration of multiple "OMIC" biomarkers: A precision medicine strategy for lung cancer. Lung Cancer 2017;107:50-8. [Crossref] [PubMed]
- Farhat FS, Houhou W. Targeted therapies in non-small cell lung carcinoma: what have we achieved so far? Ther Adv Med Oncol 2013;5:249-70. [Crossref] [PubMed]
- Khoo C, Rogers TM, Fellowes A, et al. Molecular methods for somatic mutation testing in lung adenocarcinoma: EGFR and beyond. Transl Lung Cancer Res 2015;4:126-41. [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Rykova EY, Morozkin ES, Ponomaryova AA, et al. Cell-free and cell-bound circulating nucleic acid complexes: mechanisms of generation, concentration and content. Expert Opin Biol Ther 2012;12:S141-53. [Crossref] [PubMed]
- Warton K, Samimi G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Front. Mol Biosci 2015;2:13. [Crossref] [PubMed]
- Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer--a survey. Biochim Biophys Acta 2007;1775:181-232. [PubMed]
- Radpour R, Barekati Z, Kohler C, et al. Hypermethylation of tumor suppressor genes involved in critical regulatory pathways for developing a blood-based test in breast cancer. PLoS One 2011;6:e16080 [Crossref] [PubMed]
- Warton K, Lin V, Navin T, et al. Methylation-capture and next-generation sequencing of free circulating DNA from human plasma. BMC Genomics 2014;15:476. [Crossref] [PubMed]
- Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA 2005;102:16368-73. [Crossref] [PubMed]
- Sikora K, Bedin C, Vicentini C, et al. Evaluation of cell-free DNA as a biomarker for pancreatic malignancies. Int J Biol Markers 2015;30:e136-41. [Crossref] [PubMed]
- Yu SC, Lee SW, Jiang P, et al. High-resolution profiling of fetal DNA clearance from maternal plasma by massively parallel sequencing. Clin Chem 2013;59:1228-37. [Crossref] [PubMed]
- Anker P, Lyautey J, Lefort F, et al. Transformation of NIH/3T3 cells and SW 480 cells displaying K-ras mutation. C R Acad Sci III 1994;317:869-74. [PubMed]
- Fan G, Zhang K, Ding J, et al. Prognostic value of EGFR and KRAS in circulating tumor DNA in patients with advanced non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget 2017;8:33922-32. [PubMed]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Warton K, Mahon KL, Samimi G. Methylated circulating tumor DNA in blood: power in cancer prognosis and response. Endocr Relat Cancer 2016;23:R157-71. [Crossref] [PubMed]
- Esteller M. Epigenetics in cancer. N Engl J Med 2008;358:1148-59. [Crossref] [PubMed]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007;128:683-92. [Crossref] [PubMed]
- Lu Y, Li S, Zhu S, et al. Methylated DNA/RNA in body fluids as biomarkers for lung cancer. Biol Proced Online 2017;19:2. [Crossref] [PubMed]
- Prendergast GC, Ziff EB. Methylation-sensitive Sequence-Specific DNA Binding by the c-Myc Basic Region. Science 1991;251:186-9. [Crossref] [PubMed]
- Watt F, Molloy PL. Cytosine Methylation prevents binding to DNA of a HeLa cell Transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev 1988;2:1136-43. [Crossref] [PubMed]
- Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding Protein MeCP2 involves a histone deacetylase complex. Nature 1998;393:386-9. [Crossref] [PubMed]
- Powrózek T, Krawczyk P, Nicoś M, et al. Methylation of the DCLK1 promoter region in circulating free DNA and its prognostic value in lung cancer patients. Clin Transl Oncol 2016;18:398-404. [Crossref] [PubMed]
- Tsou JA, Galler JS, Siegmund KD, et al. Identification of a panel of sensitive and specific DNA methylation markers for lung adenocarcinoma. Mol Cancer 2007;6:70. [Crossref] [PubMed]
- Suzuki M, Shiraishi K, Eguchi A, et al. Aberrant methylation of LINE-1, SLIT2, MAL and IGFBP7 in non-small cell lung cancer. Oncol Rep 2013;29:1308-14. [Crossref] [PubMed]
- Chalitchagorn K, Shuangshoti S, Hourpai N, et al. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene 2004;23:8841-6. [Crossref] [PubMed]
- Daskalos A, Nikolaidis G, Xinarianos G, et al. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer 2009;124:81-7. [Crossref] [PubMed]
- Kitkumthorn N, Tuangsintanakul T, Rattanatanyong P, et al. LINE-1 methylation in the peripheral blood mononuclear cells of cancer patients. Clin Chim Acta 2012;413:869-74. [Crossref] [PubMed]
- Zhu ZZ, Sparrow D, Hou L. Repetitive element hypomethylation in blood leukocyte DNA and cancer incidence, prevalence and mortality in elderly individuals: the Normative Aging Study. Cancer Causes Control 2011;22:437-47. [Crossref] [PubMed]
- Ikeda K, Shiraishi K, Eguchi A, et al. Long interspersed nucleotide element 1 hypomethylation is associated with poor prognosis of lung adenocarcinoma. Ann Thorac Surg 2013;96:1790-4. [Crossref] [PubMed]
- Saito K, Kawakami K, Matsumoto I, et al. Long interspersed nuclear element 1 hypomethylation is a marker of poor prognosis in stage IA non-small cell lung cancer. Clin Cancer Res 2010;16:2418-26. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nature Med 2008;14:985-90. [Crossref] [PubMed]
- Balgkouranidou I, Chimonidou M, Milaki G, et al. SOX17 promoter methylation in plasma circulating tumor DNA of patients with non-small cell lung cancer. Clin Chem Lab Med 2016;54:1385-93. [Crossref] [PubMed]
- Konecny M, Markus J, Waczulikova I, et al. The value of SHOX2 methylation test in peripheral blood samples used for the differential diagnosis of lung cancer and other lung disorders. Neoplasma 2016;63:246-53. [PubMed]
- Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett 2011;303:21-8. [Crossref] [PubMed]
- Margolin G, Petrykowska HM, Jameel N, et al. Robust Detection of DNA Hypermethylation of ZNF154 as a Pan-Cancer Locus with in Silico Modeling for Blood-based Diagnostic Development. J Mol Diagn 2016;18:283-98. [Crossref] [PubMed]
- Weiss G, Schlegel A, Kottwitz D, et al. Validation of the SHOX2/PTGER4 DNA Methylation Marker Panel for Plasma-Based Discrimination between Patients with Malignant and Nonmalignant Lung Disease. J Thorac Oncol 2017;12:77-84. [Crossref] [PubMed]
- Usadel H, Brabender J, Danenberg KD, et al. Quantitative Adenomatouspolyposiscoli Promoter Methylation Analysis in Tumor Tissue, Serum, and Plasma DNA of Patients with Lung Cancer. Cancer Res 2002;62:371-5. [PubMed]
- Hsu HS, Chen TP, Hung C, et al. Characterization of a Multiple Epigenetic Marker Panel for Lung Cancer Detection and Risk Assessment in Plasma. Cancer 2007;110:2019-26. [Crossref] [PubMed]
- Zhang Y, Miao Y, Yi J, et al. Frequent Epigenetic Inactivation of Deleted in Lung and Esophageal Cancer 1 Gene by Promoter Methylation in Non-small-cell Lung Cancer. Clin Lung Cancer 2010;11:264-70. [Crossref] [PubMed]
- Gainetdinov IV, Kapitskaya KY, Rykova EY, et al. Hypomethylation of Human-specific Family of LINE-1 Retrotransposons in Circulating DNA of Lung Cancer Patients. Lung Cancer 2016;99:127-30. [Crossref] [PubMed]
- Bearzatto A, Conte D, Frattini M, et al. p16(INK4A) Hypermethylation Detected by Fluorescent Methylation-specific PCR in Plasma from Non-small Cell Lung Cancer. Clin Cancer Res 2002;8:3782-7. [PubMed]
- An Q, Liu Y, Gao Y, et al. Detection of p16 Hypermethylation in Circulating Plasma DNA of Non-small Cell Lung Cancer Patients. Cancer Lett 2002;188:109-14. [Crossref] [PubMed]
- Xiao P, Chen JR, Zhou F, et al. Methylation of P16 in Exhaled Breath Condensate for Diagnosis of Non-small Cell Lung Cancer. Lung Cancer 2014;83:56-60. [Crossref] [PubMed]
- Ponomaryova AA, Rykova EY, Cherdyntseva NV, et al. RARβ2 Gene Methylation Level in the Circulating DNA from Blood of Patients with Lung Cancer. Eur J Cancer Prev 2011;20:453-5. [Crossref] [PubMed]
- Powrózek T, Krawczyk P, Kucharczyk T, et al. Septin 9 Promoter Region Methylation in Free Circulating DNA potential Role in Noninvasive Diagnosis of Lung Cancer: Preliminary Report. Med Oncol 2014;31:917. [Crossref] [PubMed]
- Kneip C, Schmidt B, Seegebarth A, et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J Thorac Oncol 2011;6:1632-8. [Crossref] [PubMed]
- Balgkouranidou I, Chimonidou M, Milaki G, et al. Breast cancer metastasis suppressor-1 promoter methylation in cell-free DNA provides prognostic information in non-small cell lung cancer. Br J Cancer 2014;110:2054-62. [Crossref] [PubMed]
- Ponomaryova AA, Cherdyntseva NV, Bondar AA, et al. Dynamics of LINE-1 Retrotransposon Methylation Levels in Circulating DNA from Lung Cancer Patients Undergoing Antitumor Therapy. Mol Biol (Mosk) 2017;51:622-8. [PubMed]
- Begum S, Brait M, Dasgupta S, et al. An epigenetic marker panel for detection of lung cancer using cell-free serum DNA. Clin Cancer Res 2011;17:4494-503. [Crossref] [PubMed]
- Esteller M, Sanchez-Cespedes M, Rosell R, et al. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell Lung Cancer Patients. Cancer Res 1999;59:67-70. [PubMed]
- Ramirez JL, Sarries C, de Castro PL, et al. Methylation Patterns and K-ras Mutationsin Tumor and Paired Serum of Resected Non-small Cell Lung Cancer Patients. Cancer Lett 2003;193:207-16. [Crossref] [PubMed]
- Salazar F, Molina MA, Sanchez-Ronco M, et al. Firstline therapy and methylation status of CHFR in serum influence outcome to chemotherapy versus EGFR tyrosine kinase inhibitors as second-line therapy in stage IV non-small-cell lung cancer patients. Lung Cancer 2011;72:84-91. [Crossref] [PubMed]
- Ramirez JL, Rosell R, Taron M, et al. 14-3-3sigma methylation in pretreatment serum circulating DNA of cisplatin-plus-gemcitabine-treated advanced non-small-cell lung cancer patients predicts survival: The Spanish Lung Cancer Group. J Clin Oncol 2005;23:9105-12. [Crossref] [PubMed]
- El Messaoudi S, Rolet F, Mouliere F, et al. Circulating cell free DNA: Preanalytical considerations. Clin Chim Acta 2013;424:222-30. [Crossref] [PubMed]
- Noehammer C, Pulverer W, Hassler MR, et al. Strategies for validation and testing of DNA methylation biomarkers. Epigenomics 2014;6:603-22. [Crossref] [PubMed]
- Lissa D, Robles A. Methylation analyses in liquid biopsy. Transl Lung Cancer Res 2016;5:492-504. [Crossref] [PubMed]
- Devonshire AS, Whale AS, Gutteridge A, et al. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem 2014;406:6499-512. [Crossref] [PubMed]
- Mauger F, Dulary C, Daviaud C, et al. Comprehensive evaluation of methods to isolate, quantify, and characterize circulating cell-free DNA from small volumes of plasma. Anal Bioanal Chem 2015;407:6873-8. [Crossref] [PubMed]
- Page K, Guttery DS, Zahra N. Influence of Plasma Processing on Recovery and Analysis of Circulating Nucleic Acids. PLoS One 2013;8:e77963 [Crossref] [PubMed]
- Bailey VJ, Zhang Y, Keeley BP, et al. Single-tube analysis of DNA methylation with silica superparamagnetic beads. Clin Chem 2010;56:1022-5. [Crossref] [PubMed]
- Weisenberger DJ, Trinh BN, Campan M, et al. DNA methylation analysis by digital bisulfite genomic sequencing and digital MethyLight. Nucleic Acids Res 2008;36:4689-98. [Crossref] [PubMed]
- Yu M, Carter KT, Makar KW, et al. MethyLight droplet digital PCR for detection and absolute quantification of infrequently methylated alleles. Epigenetics 2015;10:803-9. [Crossref] [PubMed]
- Li M, Chen WD, Papadopoulos N, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol 2009;27:858-63. [Crossref] [PubMed]
- Day E, Dear PH, McCaughan F. Digital PCR strategies in the development and analysis of molecular biomarkers for personalized medicine. Methods 2013;59:101-7. [Crossref] [PubMed]
- Pisanic TR, Athamanolap P, Poh W, et al. DREAMing: a simple and ultrasensitive method for assessing intratumor epigenetic heterogeneity directly from liquid biopsies. Nucleic Acids Res 2015;43:e154 [Crossref] [PubMed]
- Delaney C, Hoeltzel M, Garg SK, et al. Maternal micronutrient supplementation suppresses T cell chemokine receptor expression and function in f1 mice. J Nutr 2012;142:1329-35. [Crossref] [PubMed]
- Hayatsu H, Wataya Y, Kazushige K. The addition of sodium bisulfite to uracil and to cytosine. J Am Chem Soc 1970;92:724-6. [Crossref] [PubMed]
- Holmes EE, Jung M, Meller S, et al. Performance Evaluation of Kits for Bisulfite-Conversion of DNA from Tissues, Cell Lines, FFPE Tissues, Aspirates, Lavages, Effusions, Plasma, Serum, and Urine. PLOS ONE 2014;9:e93933 [Crossref] [PubMed]
- Huang ZH, Hu Y, Hua D, et al. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp Mol Pathol 2011;91:702-7. [Crossref] [PubMed]
- Sun FK, Fan YC, Zhao J, et al. Detection of TFPI2 methylation in the serum of hepatocellular carcinoma patients. Dig Dis Sci 2013;58:1010-15. [Crossref] [PubMed]
- Kapitskaya KY, Azhikina TL, Ponomaryova AA, et al. MIRA analysis of RARβ2 gene methylation in DNA circulating in the blood in lung cancer. Bull Exp Biol Med 2014;157:516-9. [Crossref] [PubMed]
- Hashimoto K, Kokubun S, Itoi E, et al. Improved Quantification of DNA Methylation Using Methylation-Sensitive Restriction Enzymes and Real-Time PCR. Epigenetics 2007;2:86-91. [Crossref] [PubMed]
- Liu Y, Song C, Ladas I, et al. Methylation-sensitive enrichment of minor DNA alleles using a double-strand DNA-specific nuclease. Nucleic Acids Res 2017;45:e39 [Crossref] [PubMed]
- Liloglou T, Bediaga NG, Brown BR. Epigenetic biomarkers in lung cancer. Cancer Lett 2014;342:200-12. [Crossref] [PubMed]
- Chatterjee A, Rodger EJ, Morison IM, et al. Tools and Strategies for Analysis of Genome-Wide and Gene-Specific DNA Methylation Patterns. Methods Mol Biol 2017;1537:249-77. [Crossref] [PubMed]
- Tanić M, Beck S. Epigenome-wide association studies for cancer biomarker discovery in circulating cell-free DNA: technical advances and challenges. Curr Opin Genet Dev 2017;42:48-55. [Crossref] [PubMed]
- Plongthongkum N, Diep DH, Zhang K. Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nat Rev Genet 2014;15:647-61. [Crossref] [PubMed]
- Kurdyukov S, Bullock M. DNA Methylation Analysis: Choosing the Right Method. Biology (Basel) 2016;5:E3 [Crossref] [PubMed]
- Zhai R, Zhao Y, Su L, et al. Genome-wide DNA methylation profiling of cell-free serum DNA in esophageal adenocarcinoma and Barrett esophagus. Neoplasia 2012;14:29-33. [Crossref] [PubMed]
- Miura F, Ito T. Highly sensitive targeted methylome sequencing by post-bisulfite adaptor tagging. DNA Res 2015;22:13-8. [Crossref] [PubMed]
- Chan KC, Jiang P, Chan CW. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci USA 2013;110:18761-8. [Crossref] [PubMed]
- Legendre C, Gooden GC, Johnson K, et al. Whole-genome bisulfite sequencing of cell-free DNA identifies signature associated with metastatic breast cancer. Clin Epigenetics 2015;7:100. [Crossref] [PubMed]
- Sun K, Jiang P, Chan KC, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci USA 2015;112:E5503-12. [Crossref] [PubMed]
- Wen L, Li J, Guo H, et al. Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Cell Research 2015;25:1376. [Crossref] [PubMed]
- Lehmann-Werman R, Neiman D, Zemmour H. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci U S A 2016;113:E1826-34. [Crossref] [PubMed]
- Kang S, Li Q, Chen Q, et al. CancerLocator: non-invasive cancer diagnosis and tissue-of-origin prediction using methylation profiles of cell-free DNA. Genome Biology 2017;18:53. [Crossref] [PubMed]
- Guo S, Diep D, Plongthongkum N, et al. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nature Genetics 2017;49:635-42. [Crossref] [PubMed]
- Snyder MW, Kircher M, Hill AJ, et al. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 2016;164:57-68. [Crossref] [PubMed]
- Ulz P, Thallinger GG, Auer M. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nature Genetics 2016;48:1273-8. [Crossref] [PubMed]
- Walter K, Holcomb T, Januario T, et al. Discovery and development of DNA methylation-based biomarkers for lung cancer. Epigenomics 2014;6:59-72. [Crossref] [PubMed]
- Vineis P, Chuang SC, Vaissière T, et al. DNA Methylation Changes Associated with Cancer Risk Factors and Blood Levels of Vitamin Metabolites in a Prospective Study. Epigenetics 2011;6:195-201. [Crossref] [PubMed]
- Ponomaryova AA, Rykova EY, Cherdyntseva NV, et al. Potentialities of Aberrantly Methylated Circulating DNA for Diagnostics and Post-treatment Follow-up of Lung Cancer Patients. Lung Cancer 2013;81:397-403. [Crossref] [PubMed]
- Wang W, Feng X, Duan X, et al. Establishment of two data mining models of lung cancer screening based on three gene promoter methylations combined with telomere damage. Int J Biol Markers 2017;32:e141-6. [Crossref] [PubMed]
- Belinsky SA, Grimes MG, Casas E, et al. Predicting gene promoter methylation in non-small-cell lung cancer by evaluating sputum and serum. Br J Cancer 2007;96:1278-83. [Crossref] [PubMed]
- Russo AL, Thiagalingam A, Pan H, et al. Differential DNA hypermethylation of critical genes mediates the stage-specific tobacco smoke-induced neoplastic progression of lung cancer. Clin Cancer Res 2005;11:2466-70. [Crossref] [PubMed]
- Leng S, Do K, Christin M, et al. Defining a Gene Promoter Methylation Signature in Sputum for Lung Cancer Risk Assessment. Clin Cancer Res 2012;18:3387-95. [Crossref] [PubMed]
- Suga Y, Miyajima K, Oikawa T, et al. Quantitative p16 and ESR1 methylation in the peripheral blood of patients with non-small-cell lung cancer. Oncol Rep 2008;20:1137-42. [PubMed]
- Wang H, Zhang B, Chen D, et al. Real-time monitoring efficiency and toxicity of chemotherapy in patients with advanced lung cancer. Clin Epigenetics 2015;7:119. [Crossref] [PubMed]
- Juergens RA, Wrangle J, Vendetti FP, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov 2011;1:598-607. [Crossref] [PubMed]
- Hulbert A, Jusue-Torres I, Stark A, et al. Early detection of lung cancer using DNA promoter hypermethylation in plasma and sputum. Clin Cancer Res 2017;23:1998-2005. [Crossref] [PubMed]
- Wielscher M, Vierlinger K, Kegler U, et al. Diagnostic performance of plasma DNA methylation profiles in lung cancer, pulmonary fibrosis and COPD. EBioMedicine 2015;2:929-36. [Crossref] [PubMed]
- Baglietto L, Ponzi E, Haycock P, et al. DNA methylation changes measured in pre-diagnostic peripheral blood samples are associated with smoking and lung cancer risk. Int J Cancer 2017;140:50-61. [Crossref] [PubMed]
- Gao X, Zhang Y, Breitling LP, et al. Tobacco smoking and methylation of genes related to lung cancer development. Oncotarget 2016;7:59017-28. [PubMed]
- Wang L, Aakre JA, Jiang R, et al. Methylation markers for small cell lung cancer in peripheral blood leukocyte DNA. J Thorac Oncol 2010;5:778-85. [Crossref] [PubMed]
- Hoshimoto S, Kuo CT, Chong KK, et al. AIM1 and LINE-1 epigenetic aberrations in tumor and serum relate to melanoma progression and disease outcome. J Invest Dermatol 2012;132:1689-97. [Crossref] [PubMed]
- Nagai Y, Sunami E, Yamamoto Y, et al. LINE-1 hypomethylation status of circulating cell-free DNA in plasma as a biomarker for colorectal cancer. Oncotarget 2017;8:11906-16. [Crossref] [PubMed]
- Sandoval J, Mendez-Gonzalez J, Nadal E. A prognostic DNA methylation signature for stage I non-small-cell lung cancer. J Clin Oncol 2013;31:4140-7. [Crossref] [PubMed]
- Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199-209. [Crossref] [PubMed]
- Schmidt B, Beyer J, Dietrich D, et al. Quantification of cell-free mSHOX2 Plasma DNA for therapy monitoring in advanced stage non-small cell (NSCLC) and small-cell lung cancer (SCLC) patients. PLoS One 2015;10:e0118195 [Crossref] [PubMed]
- Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med 2011;17:330-9. [Crossref] [PubMed]
- Castro M, Grau L, Puerta P, et al. Multiplexed methylation profiles of tumor suppressor genes and clinical outcome in lung cancer. J Transl Med 2010;8:86. [Crossref] [PubMed]
- Bradly DP, Gattuso P, Pool M, et al. CDKN2A (p16) promoter hypermethylation influences the outcome in young lung cancer patients. Diagn Mol Pathol 2012;21:207-13. [Crossref] [PubMed]
- Li FF, Yang Y, Wang XL, et al. Promoter methylation of DAPK gene may contribute to the pathogenesis of nonsmall cell lung cancer: a meta-analysis. Tumour Biol 2014;35:6011-20. [Crossref] [PubMed]
- Fischer JR, Ohnmacht U, Rieger N, et al. Promoter methylation of RASSF1A, RARβ and DAPK predict poor prognosis of patients with malignant mesothelioma. Lung Cancer 2006;54:109-16. [Crossref] [PubMed]
- Haldrup C, Mundbjerg K, Vestergaard EM, et al. DNA methylation signatures for prediction of biochemical recurrence after radical prostatectomy of clinically localized prostate cancer. J Clin Oncol 2013;31:3250-8. [Crossref] [PubMed]
- García-Baquero R, Puerta P, Beltran M, et al. Methylation of tumor suppressor genes in a novel panel predicts clinical outcome in paraffin-embedded bladder tumors. Tumour Biol 2014;35:5777-86. [Crossref] [PubMed]
- Hao X, Luo H, Krawczyk M, et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci U S A 2017;114:7414-9. [Crossref] [PubMed]
- BLUEPRINT consortium. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat Biotechnol 2016;34:726-37. [Crossref] [PubMed]
- Crary-Dooley FK, Tam ME, Dunaway KW, et al. A comparison of existing global DNA methylation assays to low-coverage whole-genome bisulfite sequencing for epidemiological studies. Epigenetics 2017;12:206-14. [Crossref] [PubMed]


