
Clinical relevance of liquid biopsy for cancer screening
Cancer screening
The progression of cancer to late stages without the appearance of symptoms is one of the main reasons for being among the leading causes of death worldwide. The development of an effective screening test that identifies asymptomatic individuals to assess their likelihood having the disease has a major objective to reduce morbidity or mortality in the screened population by early detection, when treatment is more successful (1). Therefore, an early detection of cancer, before a person shows any signs of illness, would increase the chances of recovery and patients’ overall survival and might help reduce cancer-related mortality.
A screening test differs from a diagnostic test by the fact that the second is used when a subject shows signs or symptoms, to determine the presence or absence of a disease, and is usually performed after a positive screening test to establish a definitive diagnosis.
The development of effective screening techniques for early detection does not exist for many types of cancers, and many have not proven effective in reducing cancer mortality. That was due to various reasons, in particular reduced sensitivity and specificity of the tests, inter and intra-tumoral heterogeneity (2-4) or epidemiological factors…
How to evaluate a screening test for cancer
In 1968, the World Health Organization (WHO) published guidelines on the principles and practice of screening for disease, which are often referred to as Wilson’s criteria (5). But with the emergence of new genomic technologies, the WHO modified these guidelines in 2008 with the new understanding as follows (6): “The screening program should respond to a recognized need, and its objectives should be defined at the outset. There should be a defined target population and scientific evidence of the screening program’s effectiveness. The program should integrate education, testing, clinical services and program management, along with quality assurance and mechanisms to minimize potential risks of screening. It should ensure informed choice, confidentiality and respect for autonomy, and promote equity and access to screening for the entire target population. The evaluation should be planned from the outset, and the overall benefits of screening should outweigh the harm”.
An ideal screening test for cancer would be able to perfectly discriminate between individuals who have or do not have the disease (1). In practice, screening tests could exhibit false positives and false negatives (7). The consequences of these results need to be carefully considered when evaluating the advantages and disadvantages of the test, and so taking into account its benefits on one hand and its risks on the other (8). When a new screening test is developed, it is regularly compared to the gold standard test, the best test available, which usually consists of a diagnostic test considered as definitive, like a biopsy for instance. However, the latter is often invasive, expensive, unpleasant, too late, or impractical to be used widely as a screening test (1). The new test would be less expensive or noninvasive for example. Its validity is translated by a high sensitivity, which is the ability of the test to identify correctly those who have that disease, and a high specificity reflecting its capacity to identify correctly those who do not have the disease (9).
In order to fully evaluate the performance of a screening test, particular attention must be paid to the tested cohorts. Indeed, a blind study, and an association of already diagnosed individuals and populations at risk are a necessity.
Conventional biomarkers and screening tests
Different screening tests are currently being used for various types of cancer. The pap smear test, for example, is used for cervical dysplasia or cervical cancer but it does not show a high sensitivity, however, it has a high specificity (10,11). Mammography is the most common test used for breast cancer screening (12,13), it has played a key role in reducing breast cancer mortality nevertheless it presents a limited sensitivity along with excessive false-positive results and the potential of overdiagnosis. Assays for serum markers, such as the tumor antigen CA 15-3 (cancer antigen 15-3) was also applied in patients with breast cancer, but showed an average sensitivity (55,6% sensitivity for a 98%specificity) (14). As a result, this marker is preferentially used for treatment response monitoring than screening and early diagnosis. The prostate-specific antigen (PSA) test could help find prostate cancer before symptoms appear (15), although a high PSA level does not always result from the presence of cancer. Low-dose computed tomographic (CT) screening is usually recommended for people with a high risk of developing lung cancer (16) but some cancers might be missed at screening and others might develop between screening and detection. For colorectal cancer (CRC), the two most common serum-based glycoprotein CRC markers, the cancer embryonic antigen (CEA) and the carbohydrate antigen 199 (CA199) are not appropriate for CRC screening due to their low sensitivity and the lack of specificity, especially for early-stage CRC [for CEA: sensitivity of 40.9–51.8% and specificity of 85.2–95% (17-19)] and are more appropriate to be used in monitoring the CRC recurrence or patients’ response to surgical or systemic therapy. In addition, stool based screening tests for CRC were developed. The Hemoccult fecal occult blood test (FOBT) has been used for a long time to aid physicians in detecting hidden blood in stool specimen as an early indication of CRC, with a sensitivity varying between 12.9% and 79.4% and a specificity of 86.7–97.7% (20). However, since this test has many drawbacks in CRC screening, the fecal immunochemical test (FIT) is more commonly used in current CRC screening thanks to its low cost with an overall sensitivity of 0.79 and overall specificity of 0.94 (20). Another is the fecal DNA test (21). This multitarget assay detected invasive cancers and adenomas with high-grade dysplasia with 40.8% sensitivity and 94.4% specificity (22). Cologuard is the first commercial Food and Drug Administration (FDA) approved [2014] fecal DNA test presenting a higher sensitivity than the FIT in CRC and polyps but a lower specificity. However, this test is somewhat expensive (23).
What is liquid biopsy?
Upon the National Cancer Institute (NCI) Dictionary of Cancer Terms liquid biopsy is “A test done on a sample of blood to look for cancer cells from a tumor that are circulating in the blood or for pieces of DNA from tumor cells that are in the blood” (24). To our point of view this definition is rather imperfect and one can be uncomfortable to associate a so complex entity such as cells regrouping molecules, intermolecular associations, organelles, compartmentalization, a closed concentration of factors or enzymes, and programming, which are the smallest unit that can live on its own and that makes up all living organisms and the tissues of the body, with cellular components such as macromolecules (DNA, RNA or microRNA) either strongly associated with proteins or encapsulated in micro-vesicles (25). The improbable association of such different biological entities may only rely on their circulating property and on the potential clinical use of the information provided by both biological sources. As indicated in the NCI Dictionary of Cancer Terms: “A liquid biopsy may be used to help find cancer at an early stage. It may also be used to help plan treatment or to find out how well treatment is working or if cancer has come back. Being able to take multiple samples of blood over time may also help doctors understand what kinds of molecular changes are taking place in a tumor.” Despite incoherence in the term, we will use in this review the term liquid biopsy in accordance with the NCI Dictionary terminology and conventionally in the literature. The term liquid biopsy therefore applies mainly in oncology mirroring the biopsy of the tumor tissue. In addition, this terminology cannot be used for circulating DNAs that are analysed in the field of prenatal diagnosis, severe/acute inflammation (sepsis), transplantation, or sports, for instance.
Liquid biopsies are not limited to the blood, though this is greatly where the research is focused. Urine, saliva or cervical fluid may also be used, as genetic information is also present in these fluids.
Circulating cell-free DNA (cfDNA)
cfDNA has emerged as a potential biomarker especially in cancer and is being widely investigated in translational and clinical research (25-27). It may present the opportunity to diagnose, monitor recurrence, and evaluate response to therapy solely through a non-invasive blood draw. Several efforts are being made in order to assess the potential of this biomarker for early cancer screening and qualitative as well as quantitative cfDNA alterations have been examined (26). But despite intensive research, few cfDNA-based tests have been translated to clinical practice. For instance, conflicting data regarding total nuclear cfDNA concentration made it hard for cfDNA-based tests to be developed and used in clinic: plasma cfDNA concentrations in cancer patients range from a few ng/mL to several thousand ng/mL, which overlaps with the concentration range for healthy individuals (27-29).
A cfDNA-based screening test must be able to distinguish between signals from non-cancer and pre-cancerous processes and the invasive malignancy in order to achieve high clinical sensitivity (30). It would also help if the test could provide information on the tissue origin which might be possible through circulating tumor DNA (ctDNA), given the distinct differences in the patterns of somatic alterations between different tumor types.
Until now, several groups worked on developing tests for the early screening of different types of cancer from a single blood analysis. We listed in Table 1 the most useful and efficient screening tests. For CRC for example, many groups have studied the screening or diagnostic relevance of different cfDNA parameters, and several reports were focused on the detection of methylated Septin9 in the plasma which was found to be significantly higher in patients with CRC than in patients with no evidence of disease (17), making it a potential biomarker for this type of cancer (33,34). The Epi proColon® (Epigenomics AG Corporation, Berlin, Germany), based on a real-time polymerase chain reaction (PCR) detection of methylated Septin9 from blood, is the only commercially available blood based DNA hypermethylation screening test for CRC (32) and is so far the best among the commercial blood-based cancer detection assays. The test discriminated between patients with CRC and healthy controls with a sensitivity of 75–81% and a specificity of 96–99%. Other studies were intended to evaluate, in the plasma, different hypermethylated DNA promoter regions and genes (37) previously found to be CRC specific. The highest area under the curves (AUCs) achieved were 0.85 for a test combining seven promotors along with age and gender (35). In addition, an age-adjusted panel of four cell-free nucleosomes was developed by Volition and it provided an AUC of 0.97 (0.87 if not age-adjusted) for the discrimination between CRC patients and healthy controls (Table 1). It showed high sensitivity for early stages (75 and 86 at 90% specificity for stages I and II, respectively). A second combination of four cf-nucleosome biomarkers provided an AUC of 0.72 for the discrimination of polyps from the healthy group (31).

Full table
The diagnostic potential of cfDNA was also examined in other types of cancer (43), and cfDNA levels were studied (38). For instance, quantitative analysis in lung cancer (Table 1) showed increased levels of cfDNA in cancer patients than in healthy individuals with approximately a value of 0.88 for areas under the summary receiver operating characteristic curves (44-47). Similar results were observed for breast cancer with 78% sensitivity and 83% specificity (53), and ovarian cancer (54) with a sensitivity of 70% and a specificity of 90%.
Circulating DNA consists not only of nuclear but mitochondrial DNA (mtDNA). Other studies have been published on the clinical significance of mtDNA levels and integrity in the peripheral blood in different types of cancer (Table 2) such as lung (57,58), breast (59), colorectal (60,61), non-Hodgkin lymphoma (62), and others (63-70). At this time, published data are discordant and it is impossible to draw any conclusion. The lack of pre-analytical and analytical studies on circulating cell-free mtDNA could explain in part this discordance, since it is poorly characterized and little is known about its structural properties.

Full table
Circulating tumor cells (CTCs)
The discovery of cells released in the bloodstream or escaping from the tumor is of primary importance and has led to intense research for about 20 years. CTCs are incredibly hard to isolate and do not always indicate genetically cancerous cells. The value of CTCs in diagnosing different types of cancers has been also assessed in several studies (Table 3) (71,72). In lung cancer for example (78), Tanaka et al. showed that CTC enumerations had an inadequate discriminating potential between patients with lung cancer and nonmalignant disease [AUC =0.598 (P=0.122)] (73). But on the other hand, other groups showed that a CTC count of more than 25 had a high sensitivity (89%) and specificity (100%) for the differentiation between benign and malignant disease (74), and a cut-off threshold of 8.7 folate receptor-positive-CTC units between the control group and patients with lung cancer presented an AUC of 0.7956 (sensitivity =77.7% and specificity =89.5%) (75). CTCs were also detected in patients with chronic obstructive pulmonary disease (COPD) (Table 3), a risk factor for lung cancer, without clinically detected lung cancer (76), in addition CTCs number was higher in patients with stage IV NSCLC compared with patients with stage IIIB (77).

Full table
While technology to capture and profile CTCs has advanced rapidly, the complexity and the weak analytical signal may limit clinical utility relative to ctDNA-based methods (38). Initial studies, such as that performed by Diaz et al., suggest that when both ctDNA and CTCs were present, ctDNA fragments outnumbered CTCs by 50 to 1 (79) providing a much higher analytical signal. Nonetheless, CTCs do not have the disadvantage of the necessity to measure very small amounts of mutated fragments in the plasma due to the important release of wild type cfDNA in some patients whose tumors are invaded by a tumor microenvironment in a large proportion (>90% of the cells). In a recent trial of lung cancer patients, ctDNA outperformed CTCs for detection of the KRAS mutation, revealing sensitivities of 96% and 52%, respectively (80). Very recently, there has been a certain enthusiasm for single cell analysis which might be of importance for screening, since this approach is technically feasible. The results on this subject are, for the moment, little discussed or not convincing (81). In addition to the paucity of CTCs’ number in blood, one of the major drawbacks of CTC analysis would be the necessity of an immediate processing (within a half day), while it would be up to 5 days for cfDNA analysis with full blood stabilizing tubes (Table 3). Nevertheless, CTCs are more related to the liquid biopsy terminology and intrinsically more powerful since determination of cellular markers may be combined to the genetic information in the same blood sample, and they are rather of relevance for real-time diagnosis of cancer progression.
Other molecular circulating biomarkers
Other circulating biomarkers were investigated for the early detection of cancer (Table 4). The diagnostic relevance of circulating cell-free microRNAs (miRNAs) was studied in the blood of patients with different types of cancer (96). A study showed that tumor-associated circulating miRNAs are elevated in the blood of breast cancer patients and associated with tumor progression (82). A multivariable signature of nine circulating miRNAs was validated and it provided a high discrimination between breast cancer patients and healthy controls with a corresponding AUC of 0.665 (83). Other circulating miRNA signatures were identified for the early diagnosis of lung malignancies (84).

Full table
Another study suggested, for breast cancer, that the presence of circulating cancer-associated macrophage like cells might have a utility as a screening tool and may differentiate patients with malignant disease, benign breast conditions, and healthy individuals (92). Tumor educated platelets (TEPs) are another studied circulating biomarker and it was shown that TEPS mRNA profiles can be used to distinguish between healthy donors and cancer patients (93,94) (Table 4).
New avenues for molecular cancer screening tests based on cfDNA analysis
Genetic alteration profile
Based on the assumption that early-detection coupled with early treatment would be key to saving lives, liquid biopsies also have the potential to allow physicians to identify patients whose tumors have specific mutations in the least invasive way possible. Several attempts were made towards this goal especially with the use of sophisticated Next-Gen sequencing methods applied on circulating DNA. Thus, the group of Velculescu recently evaluated this strategy on 138 patients with early tumors and it successfully identified the early-stage cancer in more than half of the patients using targeted error correction sequencing (TEC-Seq) (56). cfDNA analysis was used to detect the return of cancers after treatment. Authors noted that 58 genes are typically associated with breast, lung, ovarian cancer and CRC. Of the 138 cancers, they could detect 86 stage I and stage II cancers. The genes were sequenced in 100 patients and 82 of them showed the same mutations in blood samples as well as in the tumor tissue samples. None of 44 tested healthy patients as control group have cancer-derived mutations. The limitations of the study/technological strategy is the difficulty in identifying the rare DNA from cancers and in showing up results from other types of genetic alterations or mutations that a person is born with or develops during his life.
Another study showed that TP53 mutations were also detected in the plasma of 49% small cell lung cancer (SCLC) patients with significantly higher allelic fractions in cases than in controls (52).
Virus genome detection
Dennis Lo’s group very recently described an elegant study in which the strategy is to detect the Epstein-Barr virus which is involved in most nasopharyngeal cancer cases, and to hunt for viral DNA that tumors shed into the blood in large quantities, rather than rare bits of cancer cells themselves (97). Viral DNA was found in 1,112 or 5.6% of a cohort of 20,000 men. Of those, 309 also had the DNA on confirmatory tests a month later; and, 34 turned out to have cancer following endoscopy and MRI examinations. More cases were found at the earliest stage. Only one person who tested negative on screening developed nasopharyngeal cancer within a year. Clearly this approach is promising and prescription appears warranted.
Circulating DNA fragmentation
Our team first observed that (I) shorter circulating DNA molecules were more abundant in the plasma of CRC patients relative to healthy individuals; (II) the quantity of short circulating DNA fragments <145 bp is directly correlated with ctDNA concentration (98); (III) and that mutant cfDNA derived from malignant cells is highly fragmented compared to non-mutant cfDNA (99). Optimal detection by quantitative PCR (Q-PCR) of ctDNA is obtained with amplicons <100 bp (100) and Atomic Force Microscopy analysis showed that cfDNA fragments from cancer patient plasma are mostly averaging 135 bp (28). High discrimination between stage IV CRCs and healthy individuals was reported when targeting a short amplicon (63 bp) (28). In another report, we revealed that mutant cfDNA fragment proportion was much higher than non-mutant cfDNA below 145 bp size range (99). These observations were later confirmed by Leszinski et al., who showed that DNA integrity was significantly higher in patients with CRC when compared with healthy controls and with individuals with benign colorectal diseases (P=0.005 and 0.006, respectively) (101); and by Jiang et al. using massively parallel sequencing to study the size profiles of hepatocellular carcinoma patient plasma DNA samples at a single-base resolution in a genome-wide manner (102). Based upon these observations, various DNA integrity indexes were evaluated with various efficacies in discriminating healthy and cancer patients due to the lack of readily standard operating procedures (69,70), and sufficient tested patient number (98,99). Only the recent study of Tanos et al. reported statistically evaluated screening power of a specific DNA integrity index as determined by a Q-PCR method (103).
This screening strategy is based upon a differential between cfDNA structure deriving from malignant and healthy cells rather than on the cfDNA sequence. This would lead to easier implementation, and to lower screening test cost. Works on circulating DNA size profiling are ongoing to set Q-PCR and sequencing approaches toward its inclusion in a screening blood test for cancer.
A test based on circulating mtDNA: the MiTest
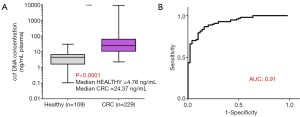
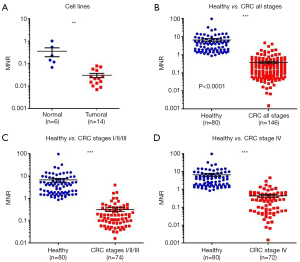
We previously showed that the amount of cfDNA may be a discriminatory factor between healthy subjects and CRC patients (28) and the receiver operating characteristic (ROC) curve analysis revealed an AUC of 0.91 (Figure 1). We also showed that quantifying and associating circulating mtDNA and nuclear DNA content enables to distinguish cancer subjects from healthy individuals (103). We determined an index based on the detection of particular sequences in the nuclear and mitochondrial genomes. When applied to cell culture supernatant, a significant difference was observed between normal and cancer cell lines (Figure 2A), and in plasma, this index is statistically lower in CRC patients than that of healthy subjects (Figure 2B,C,D). Our findings suggest that the MiTest consists of a powerful screening test for early cancer detection, and studies are ongoing to combine this marker with other parameters in order to increase the discriminative potential of the test in large cohorts of patients and healthy individuals.
Conclusions
Scientists discovered that tumors shed cells and nucleic acids into the blood circulation more than a century and 70 years ago, respectively (104,105). These molecules and cells were more recently found to reveal some of the same information that tissue biopsies provide (25,106,107). As termed here, liquid biopsy research has expanded in the last decade, generating a rapidly growing area of interest in oncology. Both academic and industry researchers from diverse areas of expertise are working on many fronts to develop, refine, and establish clinical uses for liquid biopsy tests (108).
The minimally invasive nature of liquid biopsy for malignancy without the delay, cost, and risk associated with tissue biopsy, potentially at a microscopic stage before radiologic detectability are promising advantages for cancer screening (38). Several circulating biomarkers are being investigated, from cfDNA, CTCs, circulating miRNAs and others, for the development of tests for early cancer detection. Exosomes, containing certain proteins and nucleic acids, could also be a source of multiple markers of malignancy which the analysis might be promising for the development of screening methods (109,110). But a few of these biomarkers were validated towards clinical practice. cfDNA of nuclear and mitochondrial origin seems to have an advantage in cancer screening compared to other biomarkers by showing better efficiency, and at this time, it appears to possess the characteristics to be more rapidly implemented. Combining various analysis from blood sample such as the detection by sequencing of selected mutations and genes and of protein biomarkers might be an attractive approach as very recently reported by Cohen et al. (111). While high specificity level (99%) and an overall AUC of 0.91 were observed, their data showed a moderate sensitivity (varying from 30% to 99% upon cancer types), and the cost of this multi-parametric analysis could hinder its routine use as a massive screening test. This approach should, at least, be considered for populations at risk or for specific malignant diseases.
It is to be feared or hoped that the worldwide use of a screening test will be distinguished in two ways: (I) with a test approved by public health administrations and reimbursed, followed by a statistically long and rigorous study; and (II) privately/individually (e.g., pregnancy test) with a moderate level of performance or evaluation proposed in the near future.
Standardization of the pre-analytical parameters and better knowledge on the exact origin and structure of cfDNA would provide the additional step for the implementation of its analysis. Advancement on sophisticated Q-PCR methods or Next-Gen sequencing will inevitably improve reliability of the analytical performance of the future tests. As indicated by Dennis Lo of The Chinese University of Hong Kong: “We are brick by brick putting that technology into place”. Looking forward, we may consider that liquid biopsies could add a new dimension to the cancer screening and diagnosis role of the primary care physician prior to oncology referral (38). At least, investigation of liquid biopsy screening power in tandem with other tests, such as a magnetic resonance imaging (MRI) is warranted. We envision that liquid biopsy tests may be used to screen for early-stage cancer in high-risk individuals, such as those with hereditary cancer syndromes. Nevertheless, it is crucial to further investigate these emerging biomarkers. In addition, a combined use of multiple markers may be a way to achieve more significance in early cancer detection, and increase the sensitivity and specificity of the tests. The years to come seem to be exciting, while universal screening, that constitutes the “holy grail” in oncology, appears to be accessible.
Acknowledgments
We are grateful to Safia El Messaoudi, Armèle Bonnet-Kerrache and Amaelle Otandault for their help. We would also like to thank Marc Ychou, Denis Pezet and Muriel Mathonnet for providing the blood samples.
Funding: This work was supported by the INSERM (Institut National de la Santé et de la Recherche Médicale), Lilly (France), and the SIRIC Montpellier Grant (INCa-DGOS-Inserm 6045), France.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Heidi Schwarzenbach) for the series “Technologies in Liquid Biopsies - Potential applications in Medicine” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.01.31). The series “Technologies in Liquid Biopsies - Potential applications in Medicine” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Maxim LD, Niebo R, Utell MJ. Screening tests: a review with examples. Inhal Toxicol 2014;26:811-28. [Crossref] [PubMed]
- Parsons DW, Jones S, Zhang X, et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science 2008;321:1807-12. [Crossref] [PubMed]
- Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single-cell sequencing. Nature 2011;472:90-4. [Crossref] [PubMed]
- Boesch M, Zeimet AG, Reimer D, et al. The side population of ovarian cancer cells defines a heterogeneous compartment exhibiting stem cell characteristics. Oncotarget 2014;5:7027-39. [Crossref] [PubMed]
- Wilson JM, Jungner G. Principles and practice of screening for disease. Geneva: World Health Organization, 1968.
- Andermann A, Blancquaert I, Beauchamp S, et al. Revisiting Wilson and Jungner in the genomic age: a review of screening criteria over the past 40 years. Bull World Health Organ 2008;86:317-9. [Crossref] [PubMed]
- Petticrew MP, Sowden AJ, Lister-Sharp D, et al. False-negative results in screening programmes: systematic review of impact and implications. Health Technol Assess 2000;4:1-120. [PubMed]
- PubMed Health. Benefits and risks of screening tests. 2016. Available online: https://www.ncbi.nlm.nih.gov/pubmedhealth/PMH0072602
- Hakama M, Coleman MP, Alexe DM, et al. Cancer screening: Evidence and practice in Europe 2008. Eur J Cancer 2008;44:1404-13. [Crossref] [PubMed]
- Arbyn M, Sankaranarayanan R, Muwonge R, et al. Pooled analysis of the accuracy of five cervical cancer screening tests assessed in eleven studies in Africa and India. Int J Cancer 2008;123:153-60. [Crossref] [PubMed]
- Mayrand MH, Duarte-Franco E, Rodrigues I, et al. Human Papillomavirus DNA versus Papanicolaou Screening Tests for Cervical Cancer. N Engl J Med 2007;357:1579-88. [Crossref] [PubMed]
- Friedewald SM, Rafferty EA, Rose SL, et al. Breast Cancer Screening Using Tomosynthesis in Combination With Digital Mammography. JAMA 2014;311:2499-507. [Crossref] [PubMed]
- Rafferty EA, Park JM, Philpotts LE, et al. Assessing Radiologist Performance Using Combined Digital Mammography and Breast Tomosynthesis Compared with Digital Mammography Alone: Results of a Multicenter, Multireader Trial. Radiology 2013;266:104-13. [Crossref] [PubMed]
- Stieber P, Nagel D, Blankenburg I, et al. Diagnostic efficacy of CA 15-3 and CEA in the early detection of metastatic breast cancer—A retrospective analysis of kinetics on 743 breast cancer patients. Clin Chim Acta 2015;448:228-31. [Crossref] [PubMed]
- Catalona WJ, Smith DS, Ratliff TL, et al. Measurement of Prostate-Specific Antigen in Serum as a Screening Test for Prostate Cancer. N Engl J Med 1991;324:1156-61. [Crossref] [PubMed]
- Horeweg N, Scholten ET, de Jong PA, et al. Detection of lung cancer through low-dose CT screening (NELSON): a prespecified analysis of screening test performance and interval cancers. Lancet Oncol 2014;15:1342-50. [Crossref] [PubMed]
- Tóth K, Wasserkort R, Sipos F, et al. Detection of Methylated Septin 9 in Tissue and Plasma of Colorectal Patients with Neoplasia and the Relationship to the Amount of Circulating Cell-Free DNA. PLoS One 2014;9:e115415 [Crossref] [PubMed]
- Wild N, Andres H, Rollinger W, et al. A Combination of Serum Markers for the Early Detection of Colorectal Cancer. Clin Cancer Res 2010;16:6111-21. [Crossref] [PubMed]
- Chen JS, Chen KT, Fan WC, et al. Combined analysis of survivin autoantibody and carcinoembryonic antigen biomarkers for improved detection of colorectal cancer. Clin Chem Lab Med 2010;48:719-25. [Crossref] [PubMed]
- Song LL, Li YM. Current noninvasive tests for colorectal cancer screening: An overview of colorectal cancer screening tests. World J Gastrointest Oncol 2016;8:793-800. [Crossref] [PubMed]
- Dhaliwal A, Vlachostergios PJ, Oikonomou KG, et al. Fecal DNA testing for colorectal cancer screening: Molecular targets and perspectives. World J Gastrointest Oncol 2015;7:178-83. [Crossref] [PubMed]
- Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Fecal DNA versus Fecal Occult Blood for Colorectal-Cancer Screening in an Average-Risk Population. N Engl J Med 2004;351:2704-14. [Crossref] [PubMed]
- Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget Stool DNA Testing for Colorectal-Cancer Screening. N Engl J Med 2014;370:1287-97. [Crossref] [PubMed]
- National Cancer Institute. NCI Dictionary of Cancer Terms. Available online: https://www.cancer.gov/publications/dictionaries/cancer-terms
- Thierry AR, El Messaoudi S, Gahan PB, et al. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev 2016;35:347-76. [Crossref] [PubMed]
- Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer—A survey. Biochim Biophys Acta 2007;1775:181-232. [PubMed]
- Perakis S, Auer M, Belic J, et al. Chapter Three - Advances in Circulating Tumor DNA Analysis. In: Makowski GS. Advances in Clinical Chemistry. Philadelphia: Elsevier, 2017:73-153.
- Mouliere F, El Messaoudi S, Pang D, et al. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol Oncol 2014;8:927-41. [Crossref] [PubMed]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. [Crossref] [PubMed]
- Aravanis AM, Lee M, Klausner RD. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell 2017;168:571-4. [Crossref] [PubMed]
- Rahier JF, Druez A, Faugeras L, et al. Circulating nucleosomes as new blood-based biomarkers for detection of colorectal cancer. Clin Epigenetics 2017;9:53. [Crossref] [PubMed]
- Lamb YN, Dhillon S. Epi proColon® 2.0 CE: A Blood-Based Screening Test for Colorectal Cancer. Mol Diagn Ther 2017;21:225-32. [Crossref] [PubMed]
- Jin P, Kang Q, Wang X, et al. Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. J Gastroenterol Hepatol 2015;30:830-3. [Crossref] [PubMed]
- Church TR, Wandell M, Lofton-Day C, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014;63:317-25. [Crossref] [PubMed]
- Rasmussen SL, Krarup HB, Sunesen KG, et al. Hypermethylated DNA, a circulating biomarker for colorectal cancer detection. PLoS One 2017;12:e0180809 [Crossref] [PubMed]
- Bedin C, Enzo MV, Del Bianco P, et al. Diagnostic and prognostic role of cell-free DNA testing for colorectal cancer patients. Int J Cancer 2017;140:1888-98. [Crossref] [PubMed]
- Lee BB, Lee EJ, Jung EH, et al. Aberrant Methylation of APC, MGMT, RASSF2A, and Wif-1 Genes in Plasma as a Biomarker for Early Detection of Colorectal Cancer. Clin Cancer Res 2009;15:6185-91. [Crossref] [PubMed]
- Krishnamurthy N, Spencer E, Torkamani A, et al. Liquid Biopsies for Cancer: Coming to a Patient near You. J Clin Med 2017;6:3. [Crossref] [PubMed]
- Liggett TE, Melnikov A, Yi Q, et al. Distinctive DNA methylation patterns of cell-free plasma DNA in women with malignant ovarian tumors. Gynecol Oncol 2011;120:113-20. [Crossref] [PubMed]
- Lange CP, Campan M, Hinoue T, et al. Genome-Scale Discovery of DNA-Methylation Biomarkers for Blood-Based Detection of Colorectal Cancer. PLoS One 2012;7:e50266 [Crossref] [PubMed]
- Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A 2005;102:16368-73. [Crossref] [PubMed]
- Shaw JA, Stebbing J. Circulating free DNA in the management of breast cancer. Ann Transl Med 2014;2:3. [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci Transl Med 2014;6:224ra24 [Crossref] [PubMed]
- Catarino R, Coelho A, Araújo A, et al. Circulating DNA: Diagnostic Tool and Predictive Marker for Overall Survival of NSCLC Patients. PLoS One 2012;7:e38559 [Crossref] [PubMed]
- Zhang R, Shao F, Wu X, et al. Value of quantitative analysis of circulating cell free DNA as a screening tool for lung cancer: A meta-analysis. Lung Cancer 2010;69:225-31. [Crossref] [PubMed]
- Jiang T, Zhai C, Su C, et al. The diagnostic value of circulating cell free DNA quantification in non-small cell lung cancer: A systematic review with meta-analysis. Lung Cancer 2016;100:63-70. [Crossref] [PubMed]
- Esposito A, Criscitiello C, Trapani D, et al. The Emerging Role of “Liquid Biopsies,” Circulating Tumor Cells, and Circulating Cell-Free Tumor DNA in Lung Cancer Diagnosis and Identification of Resistance Mutations. Curr Oncol Rep 2017;19:1. [Crossref] [PubMed]
- Gautschi O, Bigosch C, Huegli B, et al. Circulating Deoxyribonucleic Acid As Prognostic Marker in Non–Small-Cell Lung Cancer Patients Undergoing Chemotherapy. J Clin Oncol 2004;22:4157-64. [Crossref] [PubMed]
- Sozzi G, Conte D, Mariani L, et al. Analysis of Circulating Tumor DNA in Plasma at Diagnosis and during Follow-Up of Lung Cancer Patients. Cancer Res 2001;61:4675-8. [PubMed]
- Sozzi G, Conte D, Leon M, et al. Quantification of Free Circulating DNA As a Diagnostic Marker in Lung Cancer. J Clin Oncol 2003;21:3902-8. [Crossref] [PubMed]
- Paci M, Maramotti S, Bellesia E, et al. Circulating plasma DNA as diagnostic biomarker in non-small cell lung cancer. Lung Cancer 2009;64:92-7. [Crossref] [PubMed]
- Fernandez-Cuesta L, Perdomo S, Avogbe PH, et al. Identification of Circulating Tumor DNA for the Early Detection of Small-cell Lung Cancer. EBioMedicine 2016;10:117-23. [Crossref] [PubMed]
- Lin Z, Neiswender J, Fang B, et al. Value of circulating cell-free DNA analysis as a diagnostic tool for breast cancer: a meta-analysis. Oncotarget 2017;8:26625-36. [PubMed]
- Zhou Q, Li W, Leng B, et al. Circulating Cell Free DNA as the Diagnostic Marker for Ovarian Cancer: A Systematic Review and Meta-Analysis. PLoS One 2016;11:e0155495 [Crossref] [PubMed]
- Cohen JD, Javed AA, Thoburn C, et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc Natl Acad Sci 2017;114:10202-7. [Crossref] [PubMed]
- Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017;9:eaan2415 [Crossref] [PubMed]
- Hosgood HD, Liu CS, Rothman N, et al. Mitochondrial DNA copy number and lung cancer risk in a prospective cohort study. Carcinogenesis 2010;31:847-9. [Crossref] [PubMed]
- Meng S, De Vivo I, Liang L, et al. Pre-diagnostic leukocyte mitochondrial DNA copy number and risk of lung cancer. Oncotarget 2016;7:27307-12. [Crossref] [PubMed]
- Thyagarajan B, Wang R, Nelson H, et al. Mitochondrial DNA Copy Number Is Associated with Breast Cancer Risk. PLoS One 2013;8:e65968 [Crossref] [PubMed]
- Thyagarajan B, Wang R, Barcelo H, et al. Mitochondrial Copy Number is Associated with Colorectal Cancer Risk. Cancer Epidemiol Biomarkers Prev 2012;21:1574-81. [Crossref] [PubMed]
- Qu F, Liu X, Zhou F, et al. Association between mitochondrial DNA content in leukocytes and colorectal cancer risk. Cancer 2011;117:3148-55. [Crossref] [PubMed]
- Lan Q, Lim U, Liu CS, et al. A prospective study of mitochondrial DNA copy number and risk of non-Hodgkin lymphoma. Blood 2008;112:4247-9. [Crossref] [PubMed]
- Lu H, Busch J, Jung M, et al. Diagnostic and prognostic potential of circulating cell-free genomic and mitochondrial DNA fragments in clear cell renal cell carcinoma patients. Clin Chim Acta 2016;452:109-19. [Crossref] [PubMed]
- Fernandes J, Michel V, Camorlinga-Ponce M, et al. Circulating Mitochondrial DNA Level, a Noninvasive Biomarker for the Early Detection of Gastric Cancer. Cancer Epidemiol Biomarkers Prev 2014;23:2430-8. [Crossref] [PubMed]
- Yu M, Wan YF, Zou QH. Cell-free Circulating Mitochondrial DNA in the Serum: A Potential Non-invasive Biomarker for Ewing’s Sarcoma. Arch Med Res 2012;43:389-94. [Crossref] [PubMed]
- Li L, Hann HW, Wan S, et al. Cell-free circulating mitochondrial DNA content and risk of hepatocellular carcinoma in patients with chronic HBV infection. Sci Rep 2016;6:23992. [Crossref] [PubMed]
- Moore A, Lan Q, Hofmann JN, et al. A prospective study of mitochondrial DNA copy number and the risk of prostate cancer. Cancer Causes Control 2017;28:529-38. [Crossref] [PubMed]
- Sun Y, Zhang L, Ho SS, et al. Lower mitochondrial DNA copy number in peripheral blood leukocytes increases the risk of endometrial cancer. Mol Carcinog 2016;55:1111-7. [Crossref] [PubMed]
- Ellinger J, Albers P, Müller SC, et al. Circulating mitochondrial DNA in the serum of patients with testicular germ cell cancer as a novel noninvasive diagnostic biomarker. BJU Int 2009;104:48-52. [Crossref] [PubMed]
- Ellinger J, Müller DC, Müller SC, et al. Circulating mitochondrial DNA in serum: A universal diagnostic biomarker for patients with urological malignancies. Urol Oncol 2012;30:509-15. [Crossref] [PubMed]
- Ankeny JS, Court CM, Hou S, et al. Circulating tumour cells as a biomarker for diagnosis and staging in pancreatic cancer. Br J Cancer 2016;114:1367-75. [Crossref] [PubMed]
- Zhang Z, Fan W, Deng Q, et al. The prognostic and diagnostic value of circulating tumor cells in bladder cancer and upper tract urothelial carcinoma: a meta-analysis of 30 published studies. Oncotarget 2017;8:59527-38. [PubMed]
- Tanaka F, Yoneda K, Kondo N, et al. Circulating Tumor Cell as a Diagnostic Marker in Primary Lung Cancer. Clin Cancer Res 2009;15:6980-6. [Crossref] [PubMed]
- Fiorelli A, Accardo M, Carelli E, et al. Circulating Tumor Cells in Diagnosing Lung Cancer: Clinical and Morphologic Analysis. Ann Thorac Surg 2015;99:1899-905. [Crossref] [PubMed]
- Wang L, Wu C, Qiao L, et al. Clinical Significance of Folate Receptor-positive Circulating Tumor Cells Detected by Ligand-targeted Polymerase Chain Reaction in Lung Cancer. J Cancer 2017;8:104-10. [Crossref] [PubMed]
- Ilie M, Hofman V, Long-Mira E, et al. “Sentinel” Circulating Tumor Cells Allow Early Diagnosis of Lung Cancer in Patients with Chronic Obstructive Pulmonary Disease. PLoS One 2014;9:e111597 [Crossref] [PubMed]
- Krebs MG, Sloane R, Priest L, et al. Evaluation and Prognostic Significance of Circulating Tumor Cells in Patients With Non–Small-Cell Lung Cancer. J Clin Oncol 2011;29:1556-63. [Crossref] [PubMed]
- Hofman P. Liquid biopsy for early detection of lung cancer. Curr Opin Oncol 2017;29:73-8. [Crossref] [PubMed]
- Diaz LA, Bardelli A. Liquid Biopsies: Genotyping Circulating Tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Freidin MB, Freydina DV, Leung M, et al. Circulating Tumor DNA Outperforms Circulating Tumor Cells for KRAS Mutation Detection in Thoracic Malignancies. Clin Chem 2015;61:1299-304. [Crossref] [PubMed]
- Alix-Panabières C, Pantel K. Characterization of single circulating tumor cells. FEBS Lett 2017;591:2241-50. [Crossref] [PubMed]
- Roth C, Rack B, Müller V, et al. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res 2010;12:R90. [Crossref] [PubMed]
- Kodahl AR, Lyng MB, Binder H, et al. Novel circulating microRNA signature as a potential non-invasive multi-marker test in ER-positive early-stage breast cancer: A case control study. Mol Oncol 2014;8:874-83. [Crossref] [PubMed]
- Tomasetti M, Amati M, Neuzil J, et al. Circulating epigenetic biomarkers in lung malignancies: From early diagnosis to therapy. Lung Cancer 2017;107:65-72. [Crossref] [PubMed]
- Jiang T, Ren S, Zhou C. Role of circulating-tumor DNA analysis in non-small cell lung cancer. Lung Cancer 2015;90:128-34. [Crossref] [PubMed]
- Nikolaidis G, Raji OY, Markopoulou S, et al. DNA Methylation Biomarkers Offer Improved Diagnostic Efficiency in Lung Cancer. Cancer Res 2012;72:5692-701. [Crossref] [PubMed]
- Bearzatto A, Conte D, Frattini M, et al. p16INK4A Hypermethylation Detected by Fluorescent Methylation-specific PCR in Plasmas from Non-Small Cell Lung Cancer. Clin Cancer Res 2002;8:3782-7. [PubMed]
- Ponomaryova AA, Rykova EY, Cherdyntseva NV, et al. Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer 2013;81:397-403. [Crossref] [PubMed]
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257-61. [Crossref] [PubMed]
- Castro D, Moreira M, Gouveia AM, et al. MicroRNAs in lung cancer. Oncotarget 2017;8:81679-85. [Crossref] [PubMed]
- Sozzi G, Boeri M, Rossi M, et al. Clinical Utility of a Plasma-Based miRNA Signature Classifier Within Computed Tomography Lung Cancer Screening: A Correlative MILD Trial Study. J Clin Oncol 2014;32:768-73. [Crossref] [PubMed]
- Adams DL, Adams DK, Alpaugh RK, et al. Circulating Cancer-Associated Macrophage-Like Cells Differentiate Malignant Breast Cancer and Benign Breast Conditions. Cancer Epidemiol Biomarkers Prev 2016;25:1037-42. [Crossref] [PubMed]
- Best MG, Sol N, Kooi I, et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015;28:666-76. [Crossref] [PubMed]
- Sol N, Wurdinger T. Platelet RNA signatures for the detection of cancer. Cancer Metastasis Rev 2017;36:263-72. [Crossref] [PubMed]
- Nilsson RJ, Balaj L, Hulleman E, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood 2011;118:3680-3. [Crossref] [PubMed]
- Schwarzenbach H. Diagnostic relevance of circulating cell-free and exosomal microRNAs and long non-coding RNAs in blood of cancer patients. LaboratoriumsMedizin. 2016;40:345-53. [Crossref]
- Chan KC, Woo JK, King A, et al. Analysis of Plasma Epstein–Barr Virus DNA to Screen for Nasopharyngeal Cancer. N Engl J Med 2017;377:513-22. [Crossref] [PubMed]
- Mouliere F, Robert B, Peyrotte EA, et al. High Fragmentation Characterizes Tumour-Derived Circulating DNA. PLoS One 2011;6:e23418 [Crossref] [PubMed]
- Mouliere F, El Messaoudi S, Gongora C, et al. Circulating Cell-Free DNA from Colorectal Cancer Patients May Reveal High KRAS or BRAF Mutation Load. Transl Oncol 2013;6:319-28. [Crossref] [PubMed]
- Thierry AR, Molina F. Analytical methods for cell free nucleic acids and applications, WO/2012/028746. 2012. Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2012028746&recNum=281&docAn=EP2011065333&queryString=(%2520&
- Leszinski G, Lehner J, Gezer U, et al. Increased DNA Integrity in Colorectal Cancer. In Vivo 2014;28:299-303. [PubMed]
- Jiang P, Chan CWM, Chan KC, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci 2015;112:E1317-25. [Crossref] [PubMed]
- Thierry A, El Messaoudi S. Methods for screening a subject for cancer, WO/2016/063122. 2016. Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2016063122
- Mandel P, Metais P. Les acides nucléiques du plasma sanguin chez l’homme. C R Seances Soc Biol Fil 1948;142:241-3. [PubMed]
- Ashworth TR. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Australas Med J 1869;14:146-9.
- Stroun M, Anker P, Lyautey J, et al. Isolation and characterization of DNA from the plasma of cancer patients. Eur J Cancer Clin Oncol 1987;23:707-12. [Crossref] [PubMed]
- Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the Serum of Cancer Patients and the Effect of Therapy. Cancer Res 1977;37:646-50. [PubMed]
- National Cancer Institute. Liquid Biopsy: Using Tumor DNA in Blood to Aid Cancer Care. 2017.
- Vlassov AV, Magdaleno S, Setterquist R, et al. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta 2012;1820:940-8. [Crossref] [PubMed]
- Wang M, Ji S, Shao G, et al. Effect of exosome biomarkers for diagnosis and prognosis of breast cancer patients. Clin Transl Oncol 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926-30. [Crossref] [PubMed]


