
Cell and molecular response to IORT treatment
Introduction
Ionizing radiations (IRs), both X-rays, mainly used in conventional external beam radiotheraphy (RT), and high-energy electrons generated by intraoperative radiotherapy (IORT) linear accelerators are able to induce high stress level on either tumor or normal cells. IR causes direct or indirect damage to principal biological molecules according to its linear energy transfer (LET). When the radiation has a high LET, cell damages are mainly induced by direct ionization of macromolecules including DNA, RNA, lipids, and proteins. On the other hand, low LET radiations cause indirect damage to macromolecules, due to the generation of reactive oxygen species (ROS), especially superoxide and hydroxide radicals from the radiolysis of intracellular H2O, and reactive nitric oxide species (RNOS), which can both oxidate macromolecules and activate several intracellular signaling pathways, leading to stress responses and inflammation (1-6). Lesions involving DNA may be nitrogenous bases alterations, breaks in one (SSBs) or both DNA (DSBs) chains and chains cross-linking after breakage. Unrepaired DNA damage, due to IR can lead to mutations, genomic instability and cell death. Generally, DSBs have more lethal effects on cells than SSBs, even when induced by low LET radiation (1,3,4,7,8). In addition, although it is commonly recognized that the DNA is the principal radiation molecular target, it has recently been shown that proteins are also important IR targets that may trigger cell death mechanisms. Radiation-induced death by protein damage is thought to be caused by reduced DNA repair fidelity, indirectly reducing cell viability. There is evidence that proteins are major initial targets of free radicals and in vitro studies on cultured mammalian cell lines showed that protein oxidation may activate pro-apoptotic signaling pathways downstream of IR induced damage (9-11). In general, both DNA and protein damages contribute to the overall effect of IR toxicity, even if, which of them primarily influences cell death, has not yet been defined.
IR activates both pro- and antiproliferative signal pathways producing an imbalance in survival/apoptosis cell decision (5,6), regulated by several genes and factors involved in cell cycle progression, survival and/or cell death, DNA repair and inflammation (Figure 1). However, the contribution of these genes and signaling pathways, especially those controlling different cellular death mechanisms, need to be further investigated.
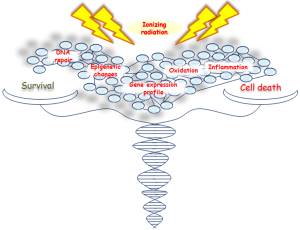
Here we describe the latest advances on cell and molecular response to IR, highlighting the most relevant research data from proteogenomic recent studies, regarding different tumor cell types including breast cancer (BC). Nowadays, the radiobiology data on high radiation doses (>10 Gy) are still very few, particularly in human cells. The possibility to clarify cell molecular strategies to choose between death and survival, after an irradiation-induced damage, opens new avenues for the selection of a proper therapy schedule, to counteract cancer growth and preserve healthy surrounding tissue from radiation effects.
Molecular response to IR: DNA repair mechanisms
Biological effects of radiation-induced cellular injury may depend on several factors. Generally, proliferating cells are more sensitive than quiescent cells to IR-induced cell death, because they have less time to repair damages (1,12-14). Cancer cell radioresistance is a complex phenomenon that may be influenced by the decrease of oxygen concentration in tumor tissue, as it is known that well-oxygenated cells are more sensitive to radiation than those with poor oxygenation. In addition, different factors, such as deregulated expression of some genes involved in cell growth, death and proliferation signalling, may influence cell radioresistance. Even if cancer cells proliferate more quickly in respect of normal cells and are more susceptible to unrepaired damage, these cells often carry multiple mutations causing constitutive activation of DNA repair mechanisms, allowing them to survive after damage, which instead would lead normal cells to death (4,15,16).
Cells have evolved complex systems to rapidly detect and efficiently repair DNA lesions, both the SSBs and DSBs (17-19). It has been observed that approximately 40 DSBs are induced for each dose delivered (1-2 Gy for most cells) and that non-transformed or non-immortalized cells, i.e., normal cells, can repair about 70 DSB/cell within 24 hours (hrs) following the radiation exposure (20,21).
Two main pathways are known for repairing DSBs: the non homologous end joining (NHEJ) and the homologous recombination (HR), that are complementary and used in different cellular conditions (Figure 2) (18,19,21). During cell cycle, these DNA repair mechanisms may be differentially activated, playing their role according to the cell cycle phase. The NHEJ drived mechanism is thought to be active during G1/G0 phases of cell cycle. Ku heterodimer is required as sensor to start NHEJ. It is formed by the Ku70 and Ku80 subunits, that recognize and bind to the DSB. Other principal factors acting are PRKDC, XRCC4, XRCC5, XRCC6, LIG4 and DCLRE1C. In proliferating cells, DSB can be repaired through a HR-dependent mechanism during the middle and late S-phase and the G2/M checkpoint requiring a homologous template. The MRN complex, formed by Mre11, Rad50 and NBN proteins, represents the DNA damages sensor, which controls the responses to DSBs via HR mechanisms. The main factors involved in HR mechanism are: RAD51, RAD52, RAD54, XRCC2 and XRCC3. Instead, DNA repair is inefficient during the S phase (22-24). Another factor that plays important roles in the cellular response to DNA damage is ATM (Ataxia Telangiectasia Mutated Protein) (25,26). It belongs to the family of phosphatidylinositol 39-kinase-like kinase (PIKK), serine/threonine protein kinases which also includes two others members, ATR (Ataxia Telangiectasia and Rad3 related) and DNA dependent protein kinase (DNA-PK).
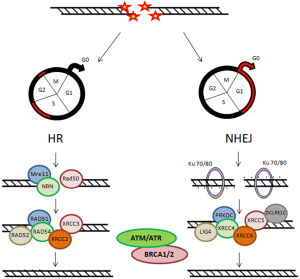
Chromatin structure is involved early, after IR injury, in particular the ATM/ATR/DNA-PK complex causes rapid phosphorylation of the histone H2AX on chromatin alongside DSBs, over some megabase of DNA regions flanking the breaks. The resulting phosphorylated H2AX (γH2AX) sites can be detected during the interphase, preferentially in euchromatic regions, by using immunofluorescence microscopy, already three minutes after IR exposure. These sites, named γH2AX foci, are also known as Ionizing Radiation Induced Foci (IRIF) (25,27,28). Afterwards γH2AX foci are formed as a platform for the recruitment or retention of other DNA repair and signaling molecules, the DNA repair processes can go beyond. Indeed, the MRN complex is rapidly localized to the γH2AX foci and the activated ATM phosphorylates Chk2, which induces Cdc25A degradation inhibiting the complexes Cdk1-Cyclin B and Cdk2-Cyclin B, with the result of cell cycle arrest (24,27). ATM also phosphorylates p53, “the genome guardian”, which exerts a crucial role following IR-induced DNA damage. In human colorectal carcinoma cell lines, the influence of p53 status on DNA damage repair after cell irradiation has been studied, applying variable IR doses until 8 Gy. It has been shown that decay of γH2AX foci is correlated with potentially lethal damage repair and p53 status, underlining that p53 functionality represents a relevant characteristic for cell survival (29). In addition, p53-binding protein 1 (53BP1) is a DNA damage response factor, classified as an adaptor/mediator required for the processing of DNA damage response signal, early recruited to damage sites and readily contributing to γH2AX foci formation. The depletion of 53BP1 results in cell cycle arrest in G2/M phase as well as in genomic instability in human and mouse cells (30,31). ATR is also recruited to DSBs sites and promotes cell cycle block through Chk proteins activation (26). The signaling via ATM/ATR can induce apoptosis or cell senescence when DNA lesions are unrepairable DSBs (24,32).
Two other important factors responsible of genomic stability maintenance, supporting efficient and precise DSB repair, are the BRCA1 and BRCA2 proteins (33). In particular, after IR exposure, BRCA1 is activated through phosphorylation by ATM and Chk2 and regulates cell-cycle arrest both during the G1-S and the G2-M checkpoints. In addition, BRCA1 has been associated with several proteins involved in the response to DNA damage and in the repair mechanism. The BRCA2 main role is to control the RAD51-mediated recombination during DSB repair by HR. The BRCA2 activity is controlled by CDKs (cyclin-dependent-kinases) in a cell cycle-dependent manner: low levels of BRCA2 phosphorylation in S phase reduce its action, while increased phosphorylation levels during G2-M progression favor the interaction with RAD51 and, therefore, the HR-mediated DNA repair mechanism (33,34). Mutations in BRCA1 and BRCA2 genes are responsible for the high risk of early onset of both hereditary breast and ovarian cancer, being hereditary BC the 20-30% of all BC cases (35-38).
Different radiation-induced cell death mechanisms
The main goal of IORT treatment, as well as radiation therapy, is to deprive cancer cells of their reproductive potential, inducing cell death to remove any remaining potential cancer cells. Nowadays, accumulating evidence reveals that induction of cell death is a very complex mechanism to account for the different therapeutic effects of IR. Indeed, in the last years it is becoming clearer that the inhibition of neoplastic cells proliferative capacity following irradiation, in particular for solid tumors, can occur through different types of cell death such as: apoptosis, necrosis, mitotic catastrophe (MC), autophagy and senescence (Figure 3) (32,39). In general, cells do not die immediately after IR treatment, but death arises after replications, frequently after 3-4 cell divisions (1,24).
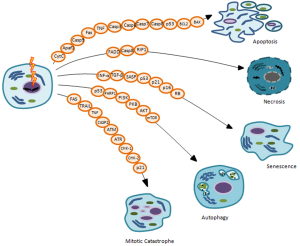
Many factors, including radiation type and dose intensity, cell type, cell cycle phase, oxygen tension, DNA repair ability, genetic variations such as Single Nucleotide Polymorphisms (SNPs) sited on genes involved in radiosensitivity and/or radiotherapy toxicity, can define the type of cell death after irradiation, briefly described below (39,40).
Apoptosis
Programed cell death or apoptosis, is a highly regulated mechanism of cell death. Distinct cytoplasmic and nuclear morphologic changes are recognizable in cells undergoing apoptosis, such as cell shrinkage, contraction and membrane blebbing, nuclear condensation, DNA fragmentation and cell destruction into membrane-bound particles (41,42). The apoptotic mechanism involves a complex network of factors according to the origin of death signal. Two principal apoptotic pathways are the well known intrinsic and extrinsic apoptosis. The intrinsic pathway is triggered by internal cell signaling, regulating mitochondrial integrity, the mitochondrial Cytochrome C release and the consequent apoptosome complex formation, composed by Apoptotic protease activating factor 1 (Apaf1) and procaspase-9. The extrinsic pathway is induced by extracellular signals transduced by the so-called transmembrane “Death Receptors” (DR, e.g., Fas with Fas Ligand), which belong to the tumor necrosis factor (TNF) receptor superfamily. Both apoptotic pathways control the activation of specific caspases, a family of cysteine-aspartic proteases involved in the apoptotic cell death mechanism. These apoptotic pathways may converge inducting the activator caspases (e.g., caspase-3, -6, -7 and -8), required for target degradation via protein lysis and DNA fragmentation (43-46).
In IR exposed cancer cells, both intrinsic and extrinsic apoptotic pathways may occur, according to delivered doses and cell type. DNA IR-induced SSBs and DSBs primarily trigger apoptosis by intrinsic pathway, when DNA lesions are unrepairable and generally via ATM/ATR signaling. Apoptotic pathways can be p53-dependent, following activation by ATM, to avoid the p53 ubiquitination by MDM2 and consequent proteosomal degradation. Moreover, p53 may be phosphorylated and activated by Chk1 and Chk2 kinase, so, it activates some pro-apoptotic proteins such as Bax, Puma, Noxa. IR-induced p53, causes a downtream activation of the death factors: Fas, Fas Ligand and KILLER/DR5 (45,47-49).
The p53 expression level and mutational status exert an important role in the cell decision to undergo death through apoptosis after irradiation. It has been observed that the tissues more sensitive to radiation-induced apoptosis, such as the spleen, the thymus and the testis, show higher levels of p53 in respect of the liver and the kidney radioresistant tissues. Tumors that result responsive to p53-dependent apoptosis are generally radiosensitive, whereas tumors that overexpress antiapoptotic proteins such as BCL2, Bcl-XL and Survivin, or do not express pro-apoptotic crucial proteins, including p53, are more radioresistant (50-53). To increase IR cancer cell apoptosis sensitivity, several specific agents, such as small molecules that structurally restore tumor-derived p53 mutant proteins, can be used to rise p53 levels (46,54). In general, different types of cancer cells, such as lung, prostate, colon cancer and immortalized keratinocytes, undergo apoptosis upon IR exposure from 1 to 20 Gy. Some non-immortalized cells show apoptotic responses only when treated with higher doses of IR (>20 Gy) (5,11,32,55).
In addition, several data show that IR treatment may induce apoptosis, p53-independent, through the membrane stress pathway with the ceramide second messenger production by sphingomyelin transmembrane signaling in vitro and in vivo (5,56).
Necrosis
Necrosis has generally been considered as a tumor cell death process that predominates after a high IR dose treatment, while at a lower dose it has been indicated as a passive and unregulated event. High radiation exposures, ranging from 32 to 50 Gy, for example, were able to induce necrosis in in vitro cultured neurons and in p53-deficient human leukemia cells. In contrast, lower IR doses, in particular 0.5 Gy of γ-rays, induced necrosis in the immortalized human keratinocyte cell line HaCaT (5,57,58). Recent studies show that IR can induce regulated cell death by necrosis in some types of tumor such as endocrine cancers, a mechanism defined as programed necrosis or necroptosis (59,60). It may be induced by apoptotic signals, particularly when the apoptotic machinery results either inefficient or blocked. Some components of the DR signaling system, such as the adaptor protein Fas-associated death domain (FADD) are common in both necrosis and apoptosis, but the final choice between these mechanisms seems to depend on caspase-8 and receptor interacting protein 1 (RIP1) activities (61,62).
In addition, it has been observed that in RIP1 expressing tumor cells, IR-induced cell death may be abolished by a small molecular inhibitor of RIP1, the necrostatin-1. On the contrary, it is possible to radiosensitize cells by increasing necroptosis using an activator of RIP1 kinase or its downstream effectors (60-62). Necrotic cells display some typical morphological characteristics, such as plasma membranes permeabilization with consequent loss of intracellular contents, organelle swelling, mitochondrial dysfunction, but unlike apoptotic, necrotic cells generally do not show any signs of DNA fragmentation (5). In contrast to apoptosis, radiation-induced necrosis is often associated with increased inflammation of the surrounding normal tissue (63).
Senescence
In normal epithelial cells, senescence is a known strategy during aging and an increase of senescent cells in older tissues or in IR treated tissues may be responsible for some pathology onsets. Several stress stimuli, in addition to IR-induced DNA damage, can trigger senescence, such as oxidative stress, chemotherapeutic agents and extended signaling by some cytokines, including interferon-α (INF-α) and transforming growth factor-β (TGF-β). Different gene expression alterations, such as deregulated expression of cell cycle regulatory proteins, which induce cell cycle arrest, upregulation of anti-apoptotic factors, high expression levels of inflammatory cytokines, growth factors and proteases, have been observed in senescent cells. These characteristics are defined as senescence associated secretory phenotype (SASP) (64-67). When grown in culture, senescent cells display a specific and typical morphology with plasma membrane and nucleus macroscopic alterations, cytoskeletal organization, changes in cell-cell interactions showing the so-called “fried egg” like appearance. A well recognized senescence marker is the senescence-associated β-galactosidase (SA-β-gal), whose increased expression has been correlated with senescence in many cell types. The DNA damage-induced signaling pathways which trigger senescence associated cell cycle arrest are mainly regulated by p53/p21 (waf1, CDKN1A), by p16 (INK4a, CDKN2A) and Rb (retinoblastoma) factors (66,67).
IR may induce accelerated cellular senescence, a state of irreversibile growth arrest in which the damaged cells show altered functions and, despite being vital, are no longer competent for proliferation. It has been demostrated that senescence is the principal response of some cell types at IR lower doses, whereas higher doses are required for the induction of apoptosis or necrosis in the same cells. In particular, a study conducted in pulmonary artery endothelial cells, irradiated with X-rays, using doses ranging from 2 to 50 Gy, has shown that increasing IR dosages induce a cell response which can change from senescence to apoptosis and/or autophagy, until necrosis at higher doses (11). Actually, most radiobiologic research papers demonstrate that there is not a unique and absolute kind of response for all cell types to a certain IR dose. For example, primary human hematopoietic cells (CD34+) undergo apoptosis whereas pulmonary artery endothelial cells become senescent when treated with the same dose of radiation (68,69).
Today the aspects establishing the specific cellular fate after IR exposure have not been clearly defined, but increasing evidence suggests that the type and radiation doses are primarly important, as well as different cell features (70).
Autophagy
Autophagy is a basic catabolic mechanism that involves cell degradation of unnecessary or dysfunctional cell components, such as damaged endoplasmic reticulum (ER) and other cytoplasmic constituents through lysosomes action. Three main different forms of autophagy have been commonly observed: microautophagy, chaperone-mediated autophagy and macroautophagy. In the context of a disease, autophagy has been described as an adaptive response to survival, whereas in other cases it appears to promote programed cell death, via non-apoptotic and caspase-independent mechanism. However, there is significant evidence that reveals a cross-talk between autophagy and apoptosis (71,72). In tumor cells undergoing chronic hypoxia and nutrient depletion, autophagy is a strategy to maintain metabolic homeostasis (73).
In normal conditions, microautophagy and chaperone-mediated autophagy permit the breakdown of abnormal proteins, cellular debris or damaged organelles, maintaining cellular homeostasis and/or as tools to recycle biological constituents (e.g., amino acid, fatty acid and energy in form of ATP). After stress stimuli, such as nutrient starvation, protein aggregation, organelle damage, oxidative or genotoxic stress, including IR, the autophagy hyper-activation promotes cell death and this case is also called macroautophagy (71-75).
A typical cell trait of autophagy is the phagophore, the site of membrane production generated when this process starts. The autophagy is mediated by protein complexes, such as class III PI3K, autophagy-related gene (Atg) proteins and other containing microtubule-associated protein 1 light-chain subunit 3 (LC3), recruited to the membrane favouring membrane expansion and phagophore elongation. Finally, the autophagosome obtained fuses with the lysosome (autophagolysosome) where hydrolases digest the cytoplasmic contents (49).
Autophagic pathways can induce survival or cell death following IR treatment, processes that might be cell and tissue specific and dependent on the expression of genes and proteins controlling apoptosis (76,77). In several types of cancer cells, such as breast, prostate, colon, lung, esophageal and glioma, IR-induced microautophagy or macroautophagy has been observed (78-83). It has been shown that following 6 Gy of IR exposure, some autophagy regulative factors significantly decreased in lung tissue, indicating a specific and strong dysregulation of IR-induced autophagy, effect not observed in liver or kidney tissues subjected to the same radiation conditions (46,84).
In the literature there is conflict with respect to the IR-triggered autophagic effect, resulting in survival or cell death promotion. Some studies show that, the autophagy preventing is radiosensitive, while the autophagy promoting is radioprotective, suggesting that IR-induced autophagy may represent an adaptive response to maintain tumor growth and survival. For example, in radioresistant BC cells a strong post-irradiation autophagy induction has been observed as a protective and pro-survival mechanism of radioresistance after exposure to IR of 4-5 Gy (85,86). In contrast with these data, other studies show that induced autophagy in some radioresistent cancer cells, including glioblastoma and lung cells, causes IR sensitization increasing cell death (84,87). In order to improve IR tumor responses, several sensitization agents to radiation-induced autophagy are currently being studied (87,88).
The molecular machinery involved in IR-induced autophagy is still not clear. IR-induced DNA damage seems to be the initiating event that causes autophagy. Recent studies show that p53 and PARP-1, a DNA repair enzyme triggered by DNA damage, exert essential roles in starting the autophagy process regulating the PI3K/PKB/AKT/mTOR signaling pathway that represents an autophagy key regulator (76,89,90).
MC
MC has initially been described as a cell death mechanism, occurring during or after aberrant mitosis, associated with various morphological and biochemical changes following radiation-induced incomplete DNA synthesis. Several evidence has revealed that it can also be caused by chemical or physical stresses and represents an oncosuppressive mechanism to avoid genomic instability. MC has been defined as a special example of apoptosis because it shows several biochemical apoptosis features, including mitochondrial membrane permeabilization and caspase activation. However, it has been observed that MC may result in death that requires both caspase-dependent and caspase-independent mechanisms (91-94). Tumor cells, harboring checkpoint deficiencies that cause incomplete DNA repair, replicative infidelity or aberrant chromosome segregation, may undergo to MC. Thus, the IR-induced loss of checkpoint controls in treated cancer cells may lead to the generation of aneuploid progeny and MC associated cell death. Cells display an increased frequency of multiple nuclei and micronuclei. In IR-treated tumor cells, MC is often associated with delayed apoptosis following increased expression of some receptors and their ligands, such as Fas, TRAIL, TNF. Moreover, caspase-2 represents the initiator caspase induced during delayed apoptosis after MC (49). Cancer cells having p53 mutations show increased IR-stimulated MC, following premature mitosis and aberrant chromosome segregation, due to high levels of cyclin B1 and frequently amplified centrosome (39,95). Generally, inhibition of factors regulating the G2/M checkpoint, such as ATM, ATR, Chk1, Chk2 and p21 favour DNA damage and, inducing aneuploidy, leads cells to MC (91,93,96).
Epigenetic changes and bystander effect IR-induced
Epigenetic modifications are heritable structural and functional genome changes occuring without changes in DNA sequence, directly affecting gene expression by mechanisms comprising histone modifications, DNA methylation and the annealing of noncoding antisense RNAs. Aberrant epigenetic events cause global changes in chromatin packaging and in specific gene promoters, influencing the transcription of genes involved in cancer development (97-99). Two principal types of changes in the DNA methylation pattern occur in cancer cells: hypo- and hyper-methylation of specific genes (100,101). It has been observed in mouse models that IR treatment with 6 Gy dose may induce effects on global hypomethylation in a sex, tissue-specific and dose-dependent manner. Most of radiation-induced epigenetic changes have been found associated with loss of methylation and decrease in expression levels of some methyltransferases, including DNMT1,DNMY3a, DNMT3b and the methyl CpG binding proteins (MeCP2) (102,103).
However, few data are available on DNA methylation changes after IR exposure in human cancer cells. In a recent study conducted on the MDA-MB-231 human BC cell line following irradiation at 2 and 6 Gy, global DNA methylation changes (at >450,000 loci) have been analized to determine potential epigenetic response to IR. The study has revealed significant differentially methylated genes related to cell cycle, DNA repair and apoptosis pathways. The degree of methylation variance of these pathways changes with radiation dose and time post-irradiation, suggesting that DNA methylation changes exert an important epigenetic role in cell response to radiation (104). In the MCF7 human BC cells treated with different fractionated IR doses (until 20 Gy), several locus-specific DNA methylation alterations have been observed, which predominantly were loss of methylation of TRAPP9, FOXC1 and LINE1 loci (105). Recently, it has been reported that radiosensitive and radioresistant cancer cells may acquire epigenetic changes at different genomic regions, in dependence of time after irradiation and cell genetic background (106). In addition, in some human colon cancer cell lines (HCT116, SW480, L174T, Co115), a relationship between enhanced cell radiation sensitivity and genomic hypomethylation induced by the DNA methyltransferase inhibitor 5-aza-cytidine (AZA) has been observed (107). Similar results were also shown by the study of Cho HJ et al. conducted on the RKO colon cancer and the MCF-7 BC cell lines, where the AZA treatment in combination with the use of the histone deacetylase (HDAC) inhibitorsodium butyrate (SB), was able to enhance radiosensitivity in both MCF-7 and RKO cell lines. The authors also noted that the combined effect caused by the demethylating agent and the HDAC inhibitor is more effective than the use of a single agent in both cancer cell lines (108). These data suggest that, the defining of specific factors regulating gene expression by DNA epigenetic changes may be a useful target for tumor radiosensitization (109,110).
Responses to IR were also observed in cells that were in contact with directly irradiated cells or have received signals from them. These responses represent the so-called non-targeted or IR “bystander effects” (111,112).
The bystander effect is increasingly considered as a long-term side effect of IR exposure. Recent studies indicate that this effect can be positive or negative and it is dependent on the radiation LET, total dose, dose rate and radiosensitivity of treated cells, similarly to the IR direct effects. The negative effects comprise apoptosis, necrosis, accelerated senescence, contributing to decreasing cell survival. In contrast, in some conditions, a positive radiation effect on bystander cells is an increased tumor cell proliferation. For example, increased cell proliferation has been observed in normal liver epithelial cells and in non-transformed fibroblasts, as well as in several transformed cells (113-115). In vitro evidence suggests that the bystander effects are communicated between cells through either the gap junction connections or by the transmission of soluble factors between irradiated cell and nonirradiated cells through the cell culture medium. Several soluble factors are involved in the bystander effect, such as ROS, nitric oxide, cyclooxygenase-2 and cytokines including TNF-α and TGF-β1 (113,116,117). In addition, an increased expression of connexin 43 in cells after 6 h of IR exposure was shown to correlate with enhanced cell to-cell communication. Nevertheless, some unanswered questions remain unclear such as the signals transmitted from irradiated to bystander cells and the relationship between the bystander response and other non-targeted effects of radiation (118).
Gene expression profile after high dose of IR
Despite the great interest of the scientific community regarding the IORT clinical application on various cancer types, a limited number of papers describe gene expression induced by IORT treatment using high doses, such as those used in IORT exclusive and in boost treatment (119), rendering the need for this field to be explored and clarified (120).
In order to highlight genes activated after high IR dose treatment, our research group has performed a gene expression profiling of BC cell lines treated with doses of 10 and 23 Gy as those doses used in BC, IORT exclusive and boost treatment, using human whole-genome microarrays. We observed consistent differences among types of treatment and cell lines (both tumorigenic and non tumorigenic). In particular, the magnitude of transcriptional variation, defined as the number of differential expressed genes is cell type and dose specific dependent (data not published). Thus, we identified candidate genes responsible for the differential cell lines response subjected to diverse doses of treatment.
Even if DNA represents the critical target for the IR biological effects, the responses generated are not solely dedicated to safe-guarding genomic integrity, but regard also the activation of critical transcription factors such as NF-κB and Activator Protein 1 (AP-1) (121,122).
NF-κB is a well-defined radiation-responsive transcription factor. Its activity modulation increases cell sensitivity in several tumor cell lines and, also, NF-κB down-regulation is probably required for TP53-dependent apoptosis. NF-κB is able to influence cell cycle regulation after irradiation and is supposed to be able to induce radioresistance by cell cycle regulation, alterations in apoptosis and changes in the ability to repair DNA damage. Disruption of NF-κB aberrant survival signaling has recently become an important issue to study therapy of several chemoresistant/radioresistant cancers (123).
AP-1 transcription factor is assembled from JUN-JUN, JUN-FOS or JUN-ATF proteins. AP-1 proteins and, above all, c-Fos play an important role in the induction and development of radiation late effects in normal tissues. JunB gene is responsive to IR and is immediately-induced early after the stimulation (124).
Moreover, the IR exposure of tumor cells induces the simultaneous compensatory activation of multiple mitogen-activated protein kinase (MAPK) pathways. These signals control survival and repopulation following radiation and this ability has been shown in various studies (125,126). It has been demonstrated that MAPK signaling is involved in cell progress through the G2-M after irradiation (125), whereas pro-survival ERK pathway is known to be activated following irradiation, in dependence on the expression of multiple growth factor receptors, autocrine factors, and Ras mutations (121,125,127).
Additional studies demonstrated that the main angiogenic regulator Vascular Endothelial Growth Factor (VEGF) is another actor supposed to be up-regulated after IR, favoring a decreased radiation sensitivity of tumor cells (128). Recently, Affolter A and colleagues described an interesting functional crosslink between VEGF and the cytoprotective MAPK-ERK in head neck squamous cell carcinoma. More precisely, they hypothesized that there might exist a feedback loop comprising VEGF-mediated activation of MAPK-ERK that in turn might cause elevated VEGF levels after cellular stress such as irradiation, suggesting that researchers targeted the ERK-VEGF axis to enhance the radiotherapy efficiency (125).
At molecular level, a number of genes have been shown to be responsive to radiation exposure. Tsai MH et al. have showed distinct differences in molecular response between a single 10 Gy high dose versus multi-fractions of 5×2 Gy dose in breast (MCF7), prostate (DU145) and glioma (SF539) cells (129). The abovementioned three cell lines responded to these types of treatments with a large comparable number of differentially expressed genes with a 1.5 or 2 fold change threshold, within a 24 hrs time course. In addition, a comparison of the time course changes in global expression patterns by multidimensional scaling analysis revealed differences rather than similarities among the cell lines, as well as between the single and multi-fractions dose regimens. More precisely, the number of genes up-regulated, by at least 2-fold, after either single or multi-fraction protocols, common to all three cell lines, was found to be small and composed by only 13 IFN-related genes. This group of genes, which are known to be transcriptionally activated by Signal Transducer and Activator Transcription 1 (STAT1), has been implicated in inflammation and may be associated with radiation resistance. The consequences of STAT1 elevation after radiation exposure could have profound effects on both normal and tumor cells.
Moreover, although p53 is one radiation-responsive gene, other genes may also contribute to the radiation response. For example, Tsai MH et al. have reported that only MCF7 cells show a cluster of p53-related genes, regulated by both single and multi-fraction schedules, while no p53-related genes were detected in either SF539 or DU145 cell lines (129). In addition, no genes were up-regulated by using the larger dose of 10 Gy, whereas there were genes predominantly up-regulated by the multi-fracionated dose in all the three cell lines. It is considered of particular interest the STAT1, up regulated in all cell lines tested, implicated in inflammation and radiation resistance. The protein encoded by this gene is a member of the STAT protein family, activated by phosphorylation in response to cytokines and growth factors, by the receptor associated kinases. Once activated, it forms homo- or heterodimers and translocate to the cell nucleus where it acts as a transcription activator. Moreover, this gene also interacts with ATM protein following DNA damage and participates in the repair of IR DNA damage (129).
Some replicated results, among array experiments involving IR high and low doses, are the induction of CDKN1A gene (120,130-140), encoding for the potent cyclin-dependent kinase inhibitor 1A (p21) and the up-regulation of GADD45 (120,131-133,137), encoding for a growth arrest and DNA-damage-inducible factor by both p53-dependent and -independent mechanisms.
Amundson SA et al., by applying Fluorescent cDNA microarray hybridization on human myeloid cell line (ML-1) assayed 4 hrs after 20 Gy IR exposure, selected 48 transcripts significantly changed by radiation treatment previously known to be radiation inducible, as well as many genes not previously reported as IR regulated. Some of these coded for proteins involved in cell cycle (such as CYCLIN B, CIP1/WAF1, GADD45 etc.), cell fate (IAP, MYC, MDM2 etc.), transcriptional regulation (JUN and FOS family members) and generally in intracellular signalling cascades that could play an important role in the induction and development of cell radiation effects (131).
Interestingly, the majority of the IR-responsive genes showed no suggestion of p53-dependent regulation. The induction of these selected stress-response genes was next measured by the authors in a panel of 12 cancer cell lines, derived from myeloid-lymphoid lineage, lung cancer, breast carcinoma and colon cancer in order to determine their role in IR-response. Particularly, only the SSAT, MBP-1, c-IAP1, RELB and BCL3 genes were primarily IR induced in all of the 12 cell lines examined (129).
Moreover, the involvement of some above described IR responsive genes was also confirmed by Jen KY and Cheung VG (132) in lymphoblastoid cells assayed at various time points within 24 hrs after irradiation, using 3 and 10 Gy. Specifically, 10 Gy induced a number of DNA repair genes, (such as factors involved in HR mechanism and previously described like RAD51C and XPC), which were not affected at the 3 Gy dose, and many cell death related genes, including a large group of anti-apoptotic and autophagy genes. In addition, the p53-regulated genes, MDM2 and PCNA, displayed increased expression levels. Following 10 Gy treatment, several MAP kinase and MAP kinase-related genes are transcriptionally induced: this signalling control survival and repopulation following radiation as previously described. Increased transcript levels of a group of oxidative stress genes were also reported in lymphoblastoid cells after 10 Gy of IR. Moreover, although some IR-responsive genes display different temporal expression patterns, depending on the dose of IR exposure, some groups of genes show very similar temporal expression patterns relative to each other at both the 3 and 10 Gy IR doses. One hundred and twenty six IR-responsive genes were in common between the two doses, including p53-dependent genes (such as CDKN1A, GADD45A, and DDB2), which play important roles in cell cycle arrest and DNA repair, general stress response genes and cell cycle-related genes (132).
Some of the above described genes are recurrent in other works, such as that published by Marko NF and colleagues (137). They analyzed the gene expression profile of human colorectal carcinoma cells treated with 20 Gy or internal beta emitter (35S-methionine), in order to compare β-radiation induced gene expression profile with that induced by external γ-radiation. Similar induction of X-IAP, IAP3, GADD45 and CDKN1A were described. In addition the authors selected a panel of 2-fold up-regulated genes only in 20 Gy IR treatment including a large number of apoptosis, transcription factors, inflammatory and degradative proteins, which may be reflective of the acute nature of the high dose. Their specific role in cell response after IR high dose has to be further highlighted. Furthermore, a comparison of temporal changes in mRNA and protein levels for p53, p21, and cdc2 showed a time lag of ~2 hrs between mRNA and corresponding protein changes (137). As described, many IR induced genes are p53 regulated but there is also a substantial p53-independent IR transcriptional response, with NF-κB playing a contributing role to radioresistance. As p53, NF-κB activates a varied set of genes ranging from cyclins to those involved in lipid signaling and translation (131,141,142). Considering that half of human cancers have a mutated p53 gene, these pathways should be further targeted to improve cancer cells radiosensitivization. The same concept also regards the GADD45 gene, up regulated after IR in numerous microarray experiments.
Microarrays solely define differences in two defined mRNA populations, a treated sample and a control, so it has been difficult to define the pure response to IR, as the control samples can be different from one experiment to another. Few experiments conducted in different laboratories were performed under similar conditions and experimental procedures. Thus, the high variability of transcriptional responses described in different cell lines emphasizes that a single cell line or cell type cannot provide a general model of response to IR stress. Finally, as technology increases in complexity, the correlations between the proteomic, phospho-proteomic and transcriptional profiles of IR treated cells will yield a more cohesive picture of cell responses to this DNA damaging stress.
Conclusions
The main goal of IORT treatments is to deprive cancer cells of their reproductive potential, addressing them to undergo cell death. IR activates complex cross-linked intracellular networks able to define cell fate capable of inducing the choice between survival and death. Indeed, in the last years it is becoming clearer that the inhibition of neoplastic cells proliferative capacity following irradiation, in particular for solid tumors, can occur through different types of cell death such as: apoptosis, necrosis, MC, autophagy and senescence.
In order to study molecular mechanisms activated by IR during IORT treatment, our research group have performed a gene expression profiling of BC cell lines treated with 10 and 23 Gy doses, using human whole-genome microarrays. We observed consistent differences among types of treatment and cell lines used, the magnitude of transcriptional variation is cell type specific and dose delivered dependent (data not published). Considering the BC complexity and heterogeneity (143-145), radioresistance/sensitivity to specific IR doses need to be directly tested on primary cells from human tumor biopsies, in order to improve personalized IR therapeutic effects.
Acknowledgments
Funding: This work was supported by FIRB/MERIT (RBNE089KHH).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Frederik Wenz and Elena Sperk) for the series “Intraoperative Radiotherapy” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2014.02.03). The series “Intraoperative Radiotherapy” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hall EJ, Giaccia AJ. eds. Radiobiology for the Radiologist, 6th ed. Philadelphia, PA, USA: Lippincott Williams & Wilkins, 2006:16-180.
- Zhao W, Diz DI, Robbins ME. Oxidative damage pathways in relation to normal tissue injury. Br J Radiol 2007;80:S23-31. [PubMed]
- Lomax ME, Folkes LK, O’Neill P. Biological consequences of radiation-induced DNA damage: relevance to radiotherapy. Clin Oncol (R Coll Radiol) 2013;25:578-85. [PubMed]
- Mladenov E, Magin S, Soni A, et al. DNA double-strand break repair as determinant of cellular radiosensitivity to killing and target in radiation therapy. Front Oncol 2013;3:113. [PubMed]
- Golden EB, Pellicciotta I, Demaria S, et al. The convergence of radiation and immunogenic cell death signaling pathways. Front Oncol 2012;2:88. [PubMed]
- Multhoff G, Radons J. Radiation, inflammation, and immune responses in cancer. Front Oncol 2012;2:58. [PubMed]
- Huang L, Snyder AR, Morgan WF. Radiation-induced genomic instability and its implications for radiation carcinogenesis. Oncogene 2003;22:5848-54. [PubMed]
- Huang L, Kim PM, Nickoloff JA, et al. Targeted and nontargeted effects of low-dose ionizing radiation on delayed genomic instability in human cells. Cancer Res 2007;67:1099-104. [PubMed]
- Du J, Gebicki JM. Proteins are major initial cell targets of hydroxyl free radicals. Int J Biochem Cell Biol 2004;36:2334-43. [PubMed]
- Shuryak I, Brenner DJ. Mechanistic analysis of the contributions of DNA and protein damage to radiation-induced cell death. Radiat Res 2012;178:17-24. [PubMed]
- Panganiban RA, Mungunsukh O, Day RM. X-irradiation induces ER stress, apoptosis, and senescence in pulmonary artery endothelial cells. Int J Radiat Biol 2013;89:656-67. [PubMed]
- Tamulevicius P, Wang M, Iliakis G. Homology-directed repair is required for the development of radioresistance during S phase: interplay between double-strand break repair and checkpoint response. Radiat Res 2007;167:1-11. [PubMed]
- Güerci AM, Dulout FN, Grillo CA, et al. Differential response of two cell lines sequentially irradiated with low X-ray doses. Int J Radiat Biol 2005;81:367-72. [PubMed]
- Ghardi M, Moreels M, Chatelain B, et al. Radiation-induced double strand breaks and subsequent apoptotic DNA fragmentation in human peripheral blood mononuclear cells. Int J Mol Med 2012;29:769-80. [PubMed]
- Moeller BJ, Richardson RA, Dewhirst MW. Hypoxia and radiotherapy: opportunities for improved outcomes in cancer treatment. Cancer Metastasis Rev 2007;26:241-8. [PubMed]
- Huber SM, Butz L, Stegen B, et al. Ionizing radiation, ion transports, and radioresistance of cancer cells. Front Physiol 2013;4:212. [PubMed]
- O’Driscoll M, Jeggo PA. The role of double-strand break repair - insights from human genetics. Nat Rev Genet 2006;7:45-54. [PubMed]
- Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 2011;25:409-33. [PubMed]
- Kakarougkas A, Jeggo P. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br J Radiol 2014; [Epub ahead of print]. [PubMed]
- Rothkamm K, Löbrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci U S A 2003;100:5057-62. [PubMed]
- Martin LM, Marples B, Coffey M, et al. DNA mismatch repair and the DNA damage response to ionizing radiation: making sense of apparently conflicting data. Cancer Treat Rev 2010;36:518-27. [PubMed]
- Tamulevicius P, Wang M, Iliakis G. Homology-directed repair is required for the development of radioresistance during S phase: interplay between double-strand break repair and checkpoint response. Radiat Res 2007;167:1-11. [PubMed]
- Escribano-Díaz C, Orthwein A, Fradet-Turcotte A, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell 2013;49:872-83. [PubMed]
- West CM, Barnett GC. Genetics and genomics of radiotherapy toxicity: towards prediction. Genome Med 2011;3:52. [PubMed]
- Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene 2007;26:7749-58. [PubMed]
- Fokas E, Prevo R, Hammond EM, et al. Targeting ATR in DNA damage response and cancer therapeutics Cancer Treat Rev 2014;40:109-17. [PubMed]
- Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 2007;26:7741-8. [PubMed]
- Cowell IG, Sunter NJ, Singh PB, et al. gammaH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS One 2007;2:e1057 [PubMed]
- Van Oorschot B, Oei AL, Nuijens AC, et al. Decay of γ-H2AX foci correlates with potentially lethal damage repair and P53 status in human colorectal carcinoma cells. Cell Mol Biol Lett 2013; [Epub ahead of print]. [PubMed]
- Schultz LB, Chehab NH, Malikzay A, et al. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol 2000;151:1381-90. [PubMed]
- Gupta A, Hunt CR, Chakraborty S, et al. Role of 53BP1 in the regulation of DNA double-strand break repair pathway choice. Radiat Res 2014;181:1-8. [PubMed]
- Surova O, Zhivotovsky B. Various modes of cell death induced by DNA damage. Oncogene 2013;32:3789-97. [PubMed]
- Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene 2006;25:5864-74. [PubMed]
- Esashi F, Christ N, Gannon J, et al. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature 2005;434:598-604. [PubMed]
- Yiannakopoulou E. Etiology of familial breast cancer with undetected BRCA1 and BRCA2 mutations: clinical implications. Cell Oncol (Dordr) 2014;37:1-8. [PubMed]
- Pothuri B. BRCA1- and BRCA2-related mutations: therapeutic implications in ovarian cancer. Ann Oncol 2013;24 Suppl 8:viii22-viii27.
- Karami F, Mehdipour P. A comprehensive focus on global spectrum of BRCA1 and BRCA2 mutations in breast cancer. Biomed Res Int 2013;2013:928562.
- Kadouri L, Sagi M, Goldberg Y, et al. Genetic predisposition to radiation induced sarcoma: possible role for BRCA and p53 mutations. Breast Cancer Res Treat 2013;140:207-11. [PubMed]
- Eriksson D, Stigbrand T. Radiation-induced cell death mechanisms. Tumour Biol 2010;31:363-72. [PubMed]
- Stewart RD, Yu VK, Georgakilas AG, et al. Effects of radiation quality and oxygen on clustered DNA lesions and cell death. Radiat Res 2011;176:587-602. [PubMed]
- Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell 2011;147:742-58. [PubMed]
- Sinha K, Das J, Pal PB, et al. Oxidative stress: the mitochondria-dependent and mitochondria-independent pathways of apoptosis. Arch Toxicol 2013;87:1157-80. [PubMed]
- Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol 2007;8:405-13. [PubMed]
- Ogura A, Oowada S, Kon Y, et al. Redox regulation in radiation-induced cytochrome c release from mitochondria of human lung carcinoma A549 cells. Cancer Lett 2009;277:64-71. [PubMed]
- Kuribayashi K, Finnberg N, Jeffers JR, et al. The relative contribution of pro-apoptotic p53-target genes in the triggering of apoptosis following DNA damage in vitro and in vivo. Cell Cycle 2011;10:2380-9. [PubMed]
- Panganiban RA, Snow AL, Day RM. Mechanisms of radiation toxicity in transformed and non-transformed cells. Int J Mol Sci 2013;14:15931-58. [PubMed]
- Pant V, Xiong S, Jackson JG, et al. The p53-Mdm2 feedback loop protects against DNA damage by inhibiting p53 activity but is dispensable for p53 stability, development, and longevity. Genes Dev 2013;27:1857-67. [PubMed]
- Chipuk JE, Bouchier-Hayes L, Kuwana T, et al. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science 2005;309:1732-5. [PubMed]
- Golden EB, Pellicciotta I, Demaria S, et al. The convergence of radiation and immunogenic cell death signaling pathways. Front Oncol 2012;2:88. [PubMed]
- Cuddihy AR, Bristow RG. The p53 protein family and radiation sensitivity: Yes or no? Cancer Metastasis Rev 2004;23:237-57. [PubMed]
- Rödel F, Hoffmann J, Distel L, et al. Survivin as a radioresistance factor, and prognostic and therapeutic target for radiotherapy in rectal cancer. Cancer Res 2005;65:4881-7. [PubMed]
- Grdina DJ, Murley JS, Miller RC, et al. A survivin-associated adaptive response in radiation therapy. Cancer Res 2013;73:4418-28. [PubMed]
- Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochim Biophys Acta 2009;1787:414-20.
- Zhu HB, Yang K, Xie YQ, et al. Silencing of mutant p53 by siRNA induces cell cycle arrest and apoptosis in human bladder cancer cells. World J Surg Oncol 2013;11:22. [PubMed]
- Han Y, Wang Y, Xu HT, et al. X-radiation induces non-small-cell lung cancer apoptosis by upregulation of Axin expression. Int J Radiat Oncol Biol Phys 2009;75:518-26. [PubMed]
- Kolesnick R, Fuks Z. Radiation and ceramide-induced apoptosis. Oncogene 2003;22:5897-906. [PubMed]
- Hotchkiss RS, Strasser A, McDunn JE, et al. Cell death. N Engl J Med 2009;361:1570-83. [PubMed]
- Jella KK, Garcia A, McClean B, et al. Cell death pathways in directly irradiated cells and cells exposed to medium from irradiated cells. Int J Radiat Biol 2013;89:182-90. [PubMed]
- Galluzzi L, Kroemer G. Necroptosis: a specialized pathway of programmed necrosis. Cell 2008;135:1161-3. [PubMed]
- Nehs MA, Lin CI, Kozono DE, et al. Necroptosis is a novel mechanism of radiation-induced cell death in anaplastic thyroid and adrenocortical cancers. Surgery 2011;150:1032-9. [PubMed]
- Vanden Berghe T, van Loo G, Saelens X, et al. Differential signaling to apoptotic and necrotic cell death by Fas-associated death domain protein FADD. J Biol Chem 2004;279:7925-33. [PubMed]
- Degterev A, Hitomi J, Germscheid M, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 2008;4:313-21. [PubMed]
- Mullins ME, Barest GD, Schaefer PW, et al. Radiation necrosis versus glioma recurrence: conventional MR imaging clues to diagnosis. AJNR Am J Neuroradiol 2005;26:1967-72. [PubMed]
- Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol 2013;75:685-705. [PubMed]
- Muller M. Cellular senescence: molecular mechanisms, in vivo significance, and redox considerations. Antioxid Redox Signal 2009;11:59-98. [PubMed]
- Tchkonia T, Zhu Y, van Deursen J, et al. Cellular senescence and the senescent secretory phenotype: therapeutic opportunities. J Clin Invest 2013;123:966-72. [PubMed]
- Tominaga K, Pereira-Smith OM. The role of chromatin reorganization in the process of cellular senescence. Curr Drug Targets 2012;13:1593-602. [PubMed]
- Xiao M, Inal CE, Parekh VI, et al. 5-Androstenediol promotes survival of gamma-irradiated human hematopoietic progenitors through induction of nuclear factor-kappaB activation and granulocyte colony-stimulating factor expression. Mol Pharmacol 2007;72:370-9. [PubMed]
- Mendonca MS, Chin-Sinex H, Dhaemers R, et al. Differential mechanisms of x-ray-induced cell death in human endothelial progenitor cells isolated from cord blood and adults. Radiat Res 2011;176:208-16. [PubMed]
- Lindsay KJ, Coates PJ, Lorimore SA, et al. The genetic basis of tissue responses to ionizing radiation. Br J Radiol 2007;80:S2-6. [PubMed]
- Dodson M, Darley-Usmar V, Zhang J. Cellular metabolic and autophagic pathways: traffic control by redox signaling. Free Radic Biol Med 2013;63:207-21. [PubMed]
- Denton D, Nicolson S, Kumar S. Cell death by autophagy: facts and apparent artefacts. Cell Death Differ 2012;19:87-95. [PubMed]
- Viganò A, Vasso M, Caretti A, et al. Protein modulation in mouse heart under acute and chronic hypoxia. Proteomics 2011;11:4202-17. [PubMed]
- Chen N, Karantza-Wadsworth V. Role and regulation of autophagy in cancer. Biochim Biophys Acta 2009;1793:1516-23.
- Wu WK, Coffelt SB, Cho CH, et al. The autophagic paradox in cancer therapy. Oncogene 2012;31:939-53. [PubMed]
- Rodriguez-Rocha H, Garcia-Garcia A, Panayiotidis MI, et al. DNA damage and autophagy. Mutat Res 2011;711:158-66. [PubMed]
- Palumbo S, Comincini S. Autophagy and ionizing radiation in tumors: the “survive or not survive” dilemma. J Cell Physiol 2013;228:1-8. [PubMed]
- Yi H, Liang B, Jia J, et al. Differential roles of miR-199a-5p in radiation-induced autophagy in breast cancer cells. FEBS Lett 2013;587:436-43. [PubMed]
- Chiu HW, Fang WH, Chen YL, et al. Monascuspiloin enhances the radiation sensitivity of human prostate cancer cells by stimulating endoplasmic reticulum stress and inducing autophagy. PLoS One 2012;7:e40462 [PubMed]
- Yu L, Tumati V, Tseng SF, et al. DAB2IP regulates autophagy in prostate cancer in response to combined treatment of radiation and a DNA-PKcs inhibitor. Neoplasia 2012;14:1203-12. [PubMed]
- Kim KW, Speirs CK, Jung DK, et al. The zinc ionophore PCI-5002 radiosensitizes non-small cell lung cancer cells by enhancing autophagic cell death. J Thorac Oncol 2011;6:1542-52. [PubMed]
- Pang XL, He G, Liu YB, et al. Endoplasmic reticulum stress sensitizes human esophageal cancer cell to radiation. World J Gastroenterol 2013;19:1736-48. [PubMed]
- Zhuang W, Qin Z, Liang Z. The role of autophagy in sensitizing malignant glioma cells to radiation therapy. Acta Biochim Biophys Sin (Shanghai) 2009;41:341-51. [PubMed]
- Chang SH, Minai-Tehrani A, Shin JY, et al. Beclin1-induced autophagy abrogates radioresistance of lung cancer cells by suppressing osteopontin. J Radiat Res 2012;53:422-32. [PubMed]
- Kim H, Bernard ME, Flickinger J, et al. The autophagy-inducing drug carbamazepine is a radiation protector and mitigator. Int J Radiat Biol 2011;87:1052-60. [PubMed]
- Chaachouay H, Ohneseit P, Toulany M, et al. Autophagy contributes to resistance of tumor cells to ionizing radiation. Radiother Oncol 2011;99:287-92. [PubMed]
- Kuwahara Y, Oikawa T, Ochiai Y, et al. Enhancement of autophagy is a potential modality for tumors refractory to radiotherapy. Cell Death Dis 2011;2:e177 [PubMed]
- Gewirtz DA. Autophagy and senescence in cancer therapy. J Cell Physiol 2014;229:6-9. [PubMed]
- Huang Q, Shen HM. To die or to live: the dual role of poly(ADP-ribose) polymerase-1 in autophagy and necrosis under oxidative stress and DNA damage. Autophagy 2010;6:1232. [PubMed]
- Li Y, Zhang J, Chen X, et al. Molecular machinery of autophagy and its implication in cancer. Am J Med Sci 2012;343:155-61. [PubMed]
- Castedo M, Perfettini JL, Roumier T, et al. Cell death by mitotic catastrophe: a molecular definition. Oncogene 2004;23:2825-37. [PubMed]
- Mansilla S, Priebe W, Portugal J. Mitotic catastrophe results in cell death by caspase-dependent and caspase-independent mechanisms. Cell Cycle 2006;5:53-60. [PubMed]
- Vakifahmetoglu H, Olsson M, Zhivotovsky B. Death through a tragedy: mitotic catastrophe. Cell Death Differ 2008;15:1153-62. [PubMed]
- Vitale I, Galluzzi L, Castedo M, et al. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol 2011;12:385-92. [PubMed]
- Ianzini F, Bertoldo A, Kosmacek EA, et al. Lack of p53 function promotes radiation-induced mitotic catastrophe in mouse embryonic fibroblast cells. Cancer Cell Int 2006;6:11. [PubMed]
- Vogel C, Hager C, Bastians H. Mechanisms of mitotic cell death induced by chemotherapy-mediated G2 checkpoint abrogation. Cancer Res 2007;67:339-45. [PubMed]
- Luce A, Courtin A, Levalois C, et al. Death receptor pathways mediate targeted and non-targeted effects of ionizing radiations in breast cancer cells. Carcinogenesis 2009;30:432-9. [PubMed]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 2008;132:567-82. [PubMed]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell 2007;128:747-62. [PubMed]
- Kouzarides T. Chromatin modifications and their function. Cell 2007;128:693-705. [PubMed]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007;128:683-92. [PubMed]
- Pogribny I, Raiche J, Slovack M, et al. Dose-dependence, sex- and tissue-specificity, and persistence of radiation-induced genomic DNA methylation changes. Biochem Biophys Res Commun 2004;320:1253-61. [PubMed]
- Raiche J, Rodriguez-Juarez R, Pogribny I, et al. Sex- and tissue-specific expression of maintenance and de novo DNA methyltransferases upon low dose X-irradiation in mice. Biochem Biophys Res Commun 2004;325:39-47. [PubMed]
- Antwih DA, Gabbara KM, Lancaster WD, et al. Radiation-induced epigenetic DNA methylation modification of radiation-response pathways. Epigenetics 2013;8:839-48. [PubMed]
- Kuhmann C, Weichenhan D, Rehli M, et al. DNA methylation changes in cells regrowing after fractioned ionizing radiation. Radiother Oncol 2011;101:116-21. [PubMed]
- Chaudhry MA, Omaruddin RA. Differential DNA methylation alterations in radiation-sensitive and -resistant cells. DNA Cell Biol 2012;31:908-16. [PubMed]
- Hofstetter B, Niemierko A, Forrer C, et al. Impact of genomic methylation on radiation sensitivity of colorectal carcinoma. Int J Radiat Oncol Biol Phys 2010;76:1512-9. [PubMed]
- Cho HJ, Kim SY, Kim KH, et al. The combination effect of sodium butyrate and 5-Aza-2'-deoxycytidine on radiosensitivity in RKO colorectal cancer and MCF-7 breast cancer cell lines. World J Surg Oncol 2009;7:49. [PubMed]
- Karagiannis TC, El-Osta A. Clinical potential of histone deacetylase inhibitors as stand alone therapeutics and in combination with other chemotherapeutics or radiotherapy for cancer. Epigenetics 2006;1:121-6. [PubMed]
- Camphausen K, Tofilon PJ. Inhibition of histone deacetylation: a strategy for tumor radiosensitization. J Clin Oncol 2007;25:4051-6. [PubMed]
- Morgan WF, Sowa MB. Effects of ionizing radiation in non irradiated cells. Proc Natl Acad Sci USA 2005;102:14127-8. [PubMed]
- Morgan WF, Sowa MB. Non-targeted bystander effects induced by ionizing radiation. Mutat Res 2007;616:159-64. [PubMed]
- Han W, Chen S, Yu KN, et al. Nitric oxide mediated DNA double strand breaks induced in proliferating bystander cells after alpha-particle irradiation. Mutat Res 2010;684:81-9. [PubMed]
- Sedelnikova OA, Nakamura A, Kovalchuk O, et al. DNA double-strand breaks form in bystander cells after microbeam irradiation of three-dimensional human tissue models. Cancer Res 2007;67:4295-302. [PubMed]
- Kim JG, Moon-Taek P, Heo K, et al. Epigenetics meets radiation biology as a new approach in cancer treatment. Int J Mol Sci 2013;14:15059-73. [PubMed]
- Kovalchuk O, Baulch JE. Epigenetic changes and nontargeted radiation effects-is there a link? Environ Mol Mutagen 2008;49:16-25. [PubMed]
- Baskar R, Balajee AS, Geard CR. Effects of low and high let radiations on bystander human lung fibroblast cell survival. Int J Radiat Biol 2007;83:551-9. [PubMed]
- Little JB. Cellular radiation effects and the bystander response. Mutat Res 2006;597:113-8. [PubMed]
- Xu QY, Gao Y, Liu Y, et al. Identification of differential gene expression profiles of radioresistant lung cancer cell line established by fractionated ionizing radiation in vitro. Chin Med J (Engl) 2008;121:1830-7. [PubMed]
- Snyder AR, Morgan WF. Gene expression profiling after irradiation: clues to understanding acute and persistent responses? Cancer Metastasis Rev 2004;23:259-68. [PubMed]
- Dent P, Yacoub A, Fisher PB, et al. MAPK pathways in radiation responses. Oncogene 2003;22:5885-96. [PubMed]
- McBride WH, Iwamoto KS, Syljuasen R, et al. The role of the ubiquitin/proteasome system in cellular responses to radiation. Oncogene 2003;22:5755-73. [PubMed]
- Chen X, Shen B, Xia L, et al. Activation of nuclear factor kappaB in radioresistance of TP53-inactive human keratinocytes. Cancer Res 2002;62:1213-21. [PubMed]
- Kajanne R, Miettinen P, Tenhunen M, et al. Transcription factor AP-1 promotes growth and radioresistance in prostate cancer cells. Int J Oncol 2009;35:1175-82. [PubMed]
- Affolter A, Fruth K, Brochhausen C, et al. Activation of mitogen-activated protein kinase extracellular signal-related kinase in head and neck squamous cell carcinomas after irradiation as part of a rescue mechanism. Head Neck 2011;33:1448-57. [PubMed]
- Zingg D, Riesterer O, Fabbro D, et al. Differential activation of the phosphatidylinositol 3-kinase/Akt survival pathway by ionizing radiation in tumor and primary endothelial cells. Cancer Res 2004;64:5398-406. [PubMed]
- Narang H, Krishna M. Mitogen-activated protein kinases: specificity of response to dose of ionizing radiation in liver. J Radiat Res 2004;45:213-20. [PubMed]
- Brieger J, Schroeder P, Gosepath J, et al. Vascular endothelial growth factor and basic fibroblast growth factor are released by squamous cell carcinoma cell lines after irradiation and increase resistance to subsequent irradiation. Int J Mol Med 2005;16:159-64. [PubMed]
- Tsai MH, Cook JA, Chandramouli GV, et al. Gene expression profiling of breast, prostate, and glioma cells following single versus fractionated doses of radiation. Cancer Res 2007;67:3845-52. [PubMed]
- Amundson SA, Do KT, Shahab S, et al. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat Res 2000;154:342-6. [PubMed]
- Amundson SA, Bittner M, Chen Y, et al. Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene 1999;18:3666-72. [PubMed]
- Jen KY, Cheung VG. Transcriptional response of lymphoblastoid cells to ionizing radiation. Genome Res 2003;13:2092-100. [PubMed]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001;98:5116-21. [PubMed]
- Stassen T, Port M, Nuyken I, et al. Radiation-induced gene expression in MCF-7 cells. Int J Radiat Biol 2003;79:319-31. [PubMed]
- Heinloth AN, Shackelford RE, Innes CL, et al. ATM-dependent and -independent gene expression changes in response to oxidative stress, gamma irradiation, and UV irradiation. Radiat Res 2003;160:273-90. [PubMed]
- Li Z, Xia L, Lee LM, et al. Effector genes altered in MCF-7 human breast cancer cells after exposure to fractionated ionizing radiation. Radiat Res 2001;155:543-53. [PubMed]
- Marko NF, Dieffenbach PB, Yan G, et al. Does metabolic radiolabeling stimulate the stress response? Gene expression profiling reveals differential cellular responses to internal beta vs. external gamma radiation. Faseb J 2003;17:1470-86. [PubMed]
- Heinloth AN, Shackelford RE, Innes CL, et al. Identification of distinct and common gene expression changes after oxidative stress and gamma and ultraviolet radiation. Mol Carcinog 2003;37:65-82. [PubMed]
- Balcer-Kubiczek EK, Zhang XF, Harrison GH, et al. Delayed expression of hpS2 and prolonged expression of CIP1/ WAF1/SDI1 in human tumor cells irradiated with X-rays, fission neutrons or 1 GeV/nucleon Fe ions. Int J Radiat Biol 1999;75:529-41. [PubMed]
- Robles AI, Bemmels NA, Foraker AB, et al. APAF-1 is a transcriptional target of p53 in DNA damage-induced apoptosis. Cancer Res 2001;61:6660-4. [PubMed]
- Park WY, Hwang CI, Im CN, et al. Identification of radiation-specific responses from gene expression profile. Oncogene 2002;21:8521-8. [PubMed]
- Chaudhry MA, Chodosh LA, McKenna WG, et al. Gene expression profile of human cells irradiated in G1 and G2 phases of cell cycle. Cancer Lett 2003;195:221-33. [PubMed]
- Bravatà V, Cammarata FP, Forte GI, et al. “Omics” of HER2-positive breast cancer. OMICS 2013;17:119-29. [PubMed]
- Bravatà V, Stefano A, Cammarata FP, et al. Genotyping analysis and 18FDG uptake in breast cancer patients: a preliminary research. J Exp Clin Cancer Res 2013;32:23. [PubMed]
- Minafra L, Norata R, Bravatà V, et al. Unmasking epithelial-mesenchymal transition in a breast cancer primary culture: a study report. BMC Res Notes 2012;5:343. [PubMed]


