
FBXW7 circular RNA repress glioma tumorigenesis
Gliomas are the most common and aggressive form of primary malignant tumors of the central nervous system (1). According to the World Health Organization (WHO) classification, gliomas are classified in four malignant grades (I to IV), based on their histological (e.g., cellularity, anaplastic, necrosis, and microvascular proliferation) and molecular (e.g., IDH1/2 mutations, 1p/19q deletions and ATRX expression) features. Glioblastomas (WHO grade IV) are the most aggressive subtype, with a mean survival of 16 months, and account for 50% of all gliomas (2).
To further understand the complex pathogenesis of glioblastoma, recent studies have amplified their focus on non-coding RNAs (ncRNAs). ncRNAs are key elements playing a central role in cellular development and homeostasis, and included several members, such as lncRNAs, circular RNAs (circRNAs) and miRNAs (3,4). Less than 2% of the total genome encodes protein-coding genes, suggesting that ncRNAs represent most of the human transcriptome, yet its role in cancer in general and gliomas in particular is poorly explored (5,6).
circRNAs were originally identified in plant viroids (7). Thereafter, other researchers found circRNA structures in yeast mitochondrial RNAs (8) and hepatitis delta virus (9). CircRNAs are a subtype of ncRNAs that are generally composed of >200 nucleotides (10). Differing from the linear RNAs, circRNAs are formed by a covalently closed loop structures that lacks 5′-3′ ends and a poly A tail (10), which render circRNAs more stable than linear RNA and more resistant to degradation by RNA exonuclease or RNase R (11,12).
circRNAs are widely expressed in eukaryotic cells and are verified to participate in regulating transcriptional and post-transcriptional gene expression (13). Moreover, previous studies have indicated that circRNAs may serve as miRNA molecular “sponges”, which play critical roles in several diseases (10). It was showed that Cdr1as/ciRS-7 affects miR-7 target gene activity by binding miR-7 as a miRNA sponge (10,13). Lately, a report showed that cir-ZNF609 may act as a sponge for miR-150-5p illustrating that cir-ZNF609 took part in the onset of Hirschsprung disease through the crosstalk with AKT3 by competing for shared miR-150-5p (14).
Some studies have associated circRNAs disruption with human tumorigenesis and may be potential biomarkers and therapeutic targets for cancer treatment (15,16). However, the overall role of circRNAs remain uncertain.
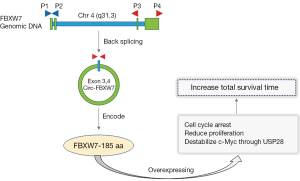
In a recent publication, Yang and co-authors from Zhang’s group from Sun Yat-Sen University, elegantly addressed the role of FBXW7 circular RNA in brain tumor (Figure 1). The authors showed that circ-FBXW7 expression was lower in glioma than normal brains and established a novel protein encoded by circ-FBXW7, FBXW7-185 aa, that suppresses glioma cell proliferation and the cell cycle. To identify this novel protein, the putative open reading frame (ORF) in circ-FBXW7 was evaluated and it was observed a potential spanning junction ORF that encoded a 185 aa protein in circ-FBXW7. This ORF is considered highly conserved among different species, implying its translation in human cells. To confirm these findings, a set of vectors was established and transfected into 293T cells. The results showed a translocation of 22 kDa protein in circ-FBXW7-transfected cells, suggesting that FBXW7-185 aa is formed by the “spanning junction ORF” (17). Then, the effects of FBXW7-185 aa on glioma cells and the molecular mechanism were evaluated and results showed that upregulation of FBXW7-185 aa in cancer cells repressed proliferation and cell cycle, while knockdown of FBXW7-185 aa promoted malignant phenotypes in vitro and in vivo (17).
FBXW7 have been shown to be strongly associated with tumorigenesis and loss of FBXW7 in cancer may have several effects on the critical cellular regulators, thereby controlling essential processes, such as cell cycle, differentiation, and apoptosis (18). Wei and colleagues also showed a decrease in FBXW7 expression level in breast cancer compared to the normal breast tissues, and the lower level of FBXW7 expression was related to shorter survival (19). Importantly, cancer genome consortiums and datasets, such as TCGA (The Cancer Genome Atlas), ICGC (International Cancer Genome Consortium) and COSMIC (Catalogue of Somatic Mutations in Cancer), highlighted the relative high frequency of FBXW7 mutation as a major cause of FBXW7 aberrant expression in cancer.
Interestingly, the Zhang’s group also observed that upregulation of FBXW7-185 aa in cancer cells repressed proliferation and cell cycle and diminished the half-life of c-Myc by antagonizing USP28-induced c-Myc stabilization. This outstanding circRNAs, FBXW7-185 aa, showed to be as “multiple safe assurances” to control cellular proliferation (17). These novel evidences are in agreement with a previous study where it was shown that FBXW7 is functioning as the tumor suppressor against several cell cycle promoters, including MYC (18). Based in these findings, Yang and colleagues have also shown that glioblastoma patients with higher circ-FBXW7 had an increased total survival time compared with those with low circ-FBXW7 expression (17).
Although Zhang’s group provides clear evidence that circRNA encodes functional protein in vivo, the clinical application of FBXW7-185 aa still has a long way to go. Hence, the clinical significance of circ-FBXW7 and FBXW7-185 aa may also need to be addressed in larger cohorts. The thorough cataloging of molecular regulatory mechanisms across the whole spectrum of gliomas affords the basis for new therapeutic development. Mackay and colleagues performed integrated molecular meta-analysis in the resource cohort of >1,000 cases of pediatric high-grade glioma and diffuse intrinsic pontine glioma. They identify co-segregating mutations in histone-mutant subgroups including loss of FBXW7 in H3.3G34R/V, and show that histone wild-type subgroups are molecularly more similar to lower-grade tumors (20). These findings greatly extended the knowledge for biological study and provided a basis for new potential therapeutic targets.
Concluding, the work published by Yang et al. in J Natl Cancer Inst have provided novel and important role of FBXW7 circular RNA in suppressing human glioma, which may have a potential prognostic implication in glioma carcinogenesis and provides the basis for novel therapeutic development. Therefore, with the improvements in our understanding of the molecular mechanisms in gliomas, it is believed that functions of several circRNAs in disease will be determined, providing evidences for the future application of circRNAs in the diagnosis, prognosis and treatment of multiple types of cancer.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Chunlin Ou (Cancer Research Institute of Central South University, Changsha, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.04.13). RMR serves as an unpaid editorial board member of Translational Cancer Research from Nov 2016 to Oct 2018. The other author has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathologica 2016;131:803-20. [Crossref] [PubMed]
- Weller M, van den Bent M, Tonn JC, et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol 2017;18:e315-29. [Crossref] [PubMed]
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Research 2014;42:D92-7. [Crossref] [PubMed]
- Park JY, Lee JE, Park JB, et al. Roles of Long Non-Coding RNAs on Tumorigenesis and Glioma Development. Brain Tumor Res Treat 2014;2:1-6. [Crossref] [PubMed]
- Sumazin P, Yang X, Chiu HS, et al. An Extensive MicroRNA-Mediated Network of RNA-RNA Interactions Regulates Established Oncogenic Pathways in Glioblastoma. Cell 2011;147:370-81. [Crossref] [PubMed]
- Chen Y, Li C, Tan C, et al. Circular RNAs: a new frontier in the study of human diseases. J Med Genet 2016;53:359-65. [Crossref] [PubMed]
- Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976;73:3852-6. [PubMed]
- Arnberg AC, Van Ommen GJ, Grivell LA, et al. Some yeast mitochondrial RNAs are circular. Cell. 1980;19:313-9. [Crossref] [PubMed]
- Kos A, Dijkema R, Arnberg AC, et al. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558-60. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733 [Crossref] [PubMed]
- Suzuki H, Zuo Y, Wang J, et al. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res 2006;34:e63 [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Peng L, Chen G, Zhu Z, et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung’s disease. Oncotarget 2017;8:808-18. [PubMed]
- Su H, Lin F, Deng X, et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med 2016;14:225. [Crossref] [PubMed]
- Song X, Zhang N, Han P, et al. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res 2016;44:e87 [Crossref] [PubMed]
- Yang Y, Gao X, Zhang M, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst 2018;110: [Crossref] [PubMed]
- Gu Z, Inomata K, Ishizawa K, et al. The FBXW7 beta-form is suppressed in human glioma cells. Biochem Biophys Res Commun 2007;354:992-8. [Crossref] [PubMed]
- Wei G, Wang Y, Zhang P, et al. Evaluating the prognostic significance of FBXW7 expression level in human breast cancer by a meta-analysis of transcriptional profiles. J Cancer Sci Ther 2012;4:299-305. [Crossref] [PubMed]
- Mackay A, Burford A, Carvalho D, et al. Integrated Molecular Meta-Analysis of 1,000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell 2017;32:520-37.e5. [Crossref] [PubMed]


