
Cyclin-dependent kinase 7 (CDK7) expression in human hepatocellular carcinoma: association with HCC progression, prognosis and cell proliferative capacity
Introduction
Hepatic cancer is one of the most common malignancies. It contributes to the sixth largest proportion of cancer mortality globally with this disease representing 9% of total cancer deaths (1). Hepatocellular carcinoma (HCC) contributes to 70–90% of total primary liver cancers, with a majority in Asia and sub-Saharan Africa (2). The multi-step progression of HCC is related to the mutations in oncogenes and the alteration of cancer suppressor genes; some of which control the cell cycle. Cyclin-dependent kinases (CDKs) mediate regular cell cycles, promoting the cell through its growth-and-division cycle (3). They maintain a regular cell cycle and arrest it when DNA damage occurs. Recent studies demonstrate that CDKs shorten the HCC cell cycle phases, enhance cell proliferation and inhibit cell apoptosis by affecting the accumulation of the transcripts of cell-cycle and anti-apoptosis signaling families (4). Such changes in the transcripts may be directly related to the increase in the malignant phenotypes of HCC.
Cyclin-dependent kinase 7 (CDK7) belongs to the CDK family, which includes serine/threonine kinases that mediate signaling pathways in the cell cycle clock. CDK7 is one of the novel CDKs, and two main mechanisms of how it takes part in cellular regulation have been characterized. First, as a dominating component of the CDK-activating kinase (CAK), CDK7 itself can phosphorylate the Thr161 site of other gatekeepers (CDK2, CDK4, and CDK6), which then drives cells to proceed from the G1 to the S phase of the cell cycle (5). Second, CDK7 is an essential competent of TFIIH, a type of transcription factor that plays a central role in the formation of the DNA repair process and the pre-initiation complex (PIC). While RNA is assembling, TFIIH phosphorylates the C-terminal domain of RNA polymerase II (POLR2A) in the early phase to initiate the elongation phase of transcription (3).
CDK7 activates other CDKs and plays a significant role in promoting cancer cell progression and proliferation (6). Both the proliferation and growth of various tumor cells are repressed when CDK7 inhibitors are applied. These cell lines include ovarian cancer, T cell acute lymphoblastic leukemia, MYCN-amplified neuroblastoma, high-grade glioma and small-cell lung cancer cell lines (7-11). Studies suggest that the inhibition of CDK7 causes a reduction in globe mRNA expression and decreases transcription factors, such as the E2F, NRF1, and CREB family, which play established roles in oncogenesis (7,11). Thus, CDK7 offers an important therapeutic target of cancer cells.
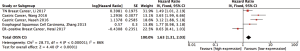
Meanwhile, a high expression of CDK7 is linked to the deteriorated prognosis of various cancers, such as gastric cancer, esophageal squamous cell carcinoma, and estrogen receptor positive and triple-negative breast cancer (6,12-15). Moreover, the overexpression of CDK7 might be positively correlated with tumor grade, stage, infiltration, and the invasion of lymph nodes in the gastric tumor (6). To investigate the influence of CDK7 expression on the tumor prognosis in general, we systematical reviewed 121 studies in the PubMed database from 2000 to July 2017. The result supported the conclusion that a high expression of CDK7 was directly related with the poor prognosis of tumors (HR =1.63, 95% CI =1.31–2.03, P<1×10−4, Figure S1). However, each individual study only reported fragmented evidence of CDK7 affecting a specific type of cancer progression and prognosis. Systematic reviews of the correlation between CDK7 expression and the pathophysiological features of cancers are still not available.
Methods
TCGA data acquisition and analysis
The data, including the CDK7 expression levels, patient phenotypes and their matched survival, were acquired from the TCGA database up until July 2017. These data were collected from the cBio Cancer Genomics Portal (www.cbioportal.org), and all the quantification files were checked and downloaded with “.txt” format.
The cutoff CDK7 expression level was defined by the mean expression. Values above the mean expression value were characterized as “high expression”, while those under this value were defined as “low expression”.
The obtained TCGA data were sorted, quantified and analyzed by SPSS 22.0 (IBM, Chicago, IL, USA). The median expression was used as the cutoff value in the Kaplan-Meier curve and Cox regression model. The Kaplan-Meier survival curve was plotted, and a log-rank test was performed to test the significance of the two different expression groups. These clinicopathological factors include the age at the initial pathological diagnosis, albumin, creatinine and fetoprotein values, fibrosis (Ishak score), sex, pathologic TNM stages and prothrombin time.
Meta-analysis of the relationship between CDK7 expression and cancer prognosis
A total of 121 studies were systematically reviewed based on the following keywords in PubMed: “(CDK7 OR “Cyclin-dependent kinase 7”) AND (cancer OR carcinoma OR tumor OR tumors OR neoplasm OR neoplasms) AND ((survival OR survivals) OR (prognosis OR prognostic OR predictive value OR progression OR progressions) OR (outcome OR follow-up OR risk)).” The search ended in July 2017. Four studies related to gastrointestinal malignancies with patient survival data were obtained for the analysis. The hazard ratio (HR) was extracted or calculated from the Kaplan-Meier curves using Engauge Digitizer 4.1 (http://digitizer.sourceforge.net).
The analysis was conducted using Review Manager 5.3 (Cochrane Collaboration, Oxford, UK), and the data were based on the qualified studies in the PubMed and TCGA databases. The correlation of CDK7 expression level and HCC prognosis was measured by the pooled HR with 95% CI. When I2>50%, a random effect model was applied, or a fixed effect model was fit otherwise.
DEGs and individual gene meta-analysis
The interactive web tool GEO2R, which allows users to compare two or more groups of samples in a GEO Series, was used to identify the DEGs from the eight independent GEO datasets. GEO2R performs comparisons on the original Series Matrix data file that was submitter-supplied using the GEOquery and limma R packages from the Bioconductor project (16,17). For the TCGA dataset, the limma package was used to identify the DEGs between the cancer and non-tumor tissue. The analysis results, including ID, P value, t, B, and logFC, are presented as tables of genes ordered by significance.
Two meta-analysis approaches were used for further analysis (18,19). We used the first method to obtain the effect size from each dataset and combined them into a meta-effect size to estimate the extent of differential expression among all the datasets. We used Hedges’ adjusted g to compute the effect size for each gene. The study-specific effect sizes were combined to obtain the pooled effect size and its standard error using the random effects inverse-variance technique. We used the second nonparametric method to conduct combination P values based on the Z-scores for each gene in each dataset (20).
Cell culture and treatment
Human HCC cell lines, including HepG2 and Skhep-1 cells, were purchased from the American Type Culture Collection (Manassas, VA, USA). The Huh7 and HCC-LM3 cells were obtained from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The Hep3B, Skhep-1, Huh7, HepG2, HCC-LM3, SMCC-7721, BEL-7402, and MHCC-97L cells were cultured in DMEM Medium (Invitrogen) supplemented with 10% fetal bovine serum (Gibco) at 37 °C in a humidified atmosphere with 5% CO2.
Plasmids construction and transfection
The CDK7 (GenBank Accession No. NM_001799) was isolated from the human cDNA library. The shRNA sequences targeting the CDK7 were as follows: shCDK7 1: CCGGCATTTAAGAGTTTCCCTGGAACTCGAGTTCCAGGGAAACTCTTAAATGTTTTT; shCDK7 2: CCGGGAAACTGATCTAGAGGTTATACTCGAGTATAACCTCTAGATCAGTTTCTTTTTTG; and shCDK7 3: CCGGGTAAATGCTGTAGAAGTGAGTCTCGAGACTCACTTCTACAGCATTTACTTTTTTG. The positive control shRNA sequence was CCGGGCCAATCGTTCTCTGACAGAACTCGAGTTCTGTCAGAGAACGATTGGCTTTTT.
For the preparation of cell transfection, the HCC SMMC-7721 and Huh7 cells were transferred from the cell culture flask to a density of 5×103 cells in 24-well plates in DMEM with 10% (v/v) FBS for 24 hr. The cells were 70–90% confluent when the transfection was initiated. A scrambled shRNA was used as the negative control. The CDK7 specific shRNAs (shCDK7), shCDK7 or the scrambled shRNA was transfected into the SMMC-7721 and Huh7 cells with the Lipofectamine 3000 Reagent (Life Technologies, USA) according to manufacturer’s protocol.
Cell proliferation assay
The bromodeoxyuridine (BrdU) assay was performed according to manufacturer’s protocol in the BrdU colorimetric cell proliferation assay kit instruction manual (cat# 1 647 229; Roche). For all the experimental conditions, the HCC SMMC-7721 and Huh7 cells were plated at a density of 1×106 cells per well in 6-well plates and were rendered quiescent by serum starvation for 36 hr. The cells were then re-stimulated with 10% (v/v) FBS for 18 hr before labeling the cells with BrdU for 6 hours. S-phase cells were visualized by microscopy and were quantitated by counting three fields of 100 cells in quadruplicate. The data are presented as the percentage of BrdU positive cells out of the 100 cells counted.
RNA purification and quantitative RT-PCR
Total RNA was extracted with the TRIzol reagent (Invitrogen). A total of 1 µg of total RNA was used for reverse transcription using the PrimeScript RT reagent kit with gDNA Eraser (Takara) according to the manufacturer’s instructions. Quantitative RT-PCR (qPCR) was performed with SYBR Green (Applied Biosystems) using the ViiA7 Real-Time PCR System (Applied Biosystems). All the qPCR reactions were performed in triplicate. The relative mRNA expression was calculated by the comparative Ct method using GAPDH as a control. The primers used in the qRT-PCR were CDK7 FW: 5'-GCAAAGCGTTATGAGAAG-3' and CDK7 REV: 5'-AAAGCATCAAGGAGACCA-3'. The data were normalized to the housekeeping gene GAPDH. The primers for housekeep gene were as follows: GAPDH FW: 5'-TGACTTCAACAGCGACACCCA-3' and GAPDH REV: 5'-CACCCTGTTGCTGTAGCCAAA-3'.
Cell viability assay with THZ1 treatment
The effects of the CDK7 inhibitor THZ1 on HCC SMMC-7721 and Huh7 cellular viability were determined by a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay using a commercial kit (Promega, China). Briefly, THZ1 was dissolved in dimethyl sulfoxide (DMSO), and the final concentration of DMSO was adjusted to 0.1%. The cells were incubated with DMSO or THZ1 for 48 h and were quantitated by an absorbance measurement at 490 nm using a microplate reader. The results are shown as the ratio of the figures tested to the untreated control and are expressed as the mean values ± standard deviation for three independent experiments.
Results
Meta-analysis of the correlation between CDK7 expression and cancer prognosis
We acquired the transcriptome and survival data from 23 types of tumors in TCGA the database to explore the association between the over-expression of CDK7 and clinical tumor prognosis. Thirteen tumor groups, with each group having a sample size of more than 100, were involved in the following research, with a total sample size of 3,887 individual patients, as shown in Figure 1A. The results showed that a high expression level of CDK7 was overall significantly associated with a poor prognosis in tumors (HR =1.15, 95% CI =1.03–1.28, P=1×10−2). Although the survival was comparable between patients with high or low CDK7 expression in the subgroup analysis of cancer in the digestive system (HR =1.19, 95% CI =0.98–1.45, P=9×10−2), we noticed that HCC prognosis was highly negatively related to the expression level of CDK7 (HR =1.51, 95% CI =1.06–2.15).
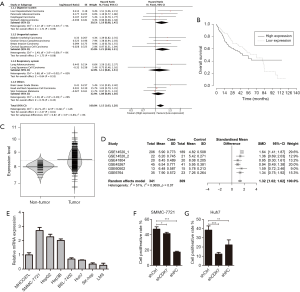
The relationship between the CDK7 expression level and patient survival
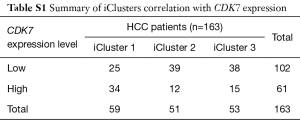
The transcriptome and survival data were combined to further explore the connection of CDK7 and HCC prognosis in HCC. There were 256 patients included in this analysis, and the cutoff value of the CDK7 expression level was defined by the mean expression. As a result, 138 patients showed a high expression of CDK7, and 118 patients showed a low expression. This proved that highly expressed CDK7 was indeed proportionally related to the poor survival time (P=2.38×10−3, Figure 1B), suggesting that CDK7 might be an important prognostic factor for overall survival in patients with HCC. In addition, we inquired about the correlation of the expression level of CDK7 and the clinicopathological factors in HCC (Table 1). The data implied that high CDK7 expression was related to neoplasm histologic grade (P<1×10−4). Multi-platform integrative molecular subtyping, performed by The Cancer Genome Atlas Research Network, divides HCC patients with different pathological and molecular features into three major groups (iCluster1, iCluster2 and iCluster 3). iCluster 1 is directly associated with the aberrant activation of cell proliferation (21,22). We found that patients with high CDK7 expression were more likely to be resolved into iCluster1 after combining the HCC iCluster data and the expression data of CDK7 (Table S1, P<1×10−3). This result may suggest that a high expression of CDK7 is positively associated with the activation of cell proliferation. Together, these findings indicate that there are significant correlations between CDK7 expression and the clinical features of HCC.
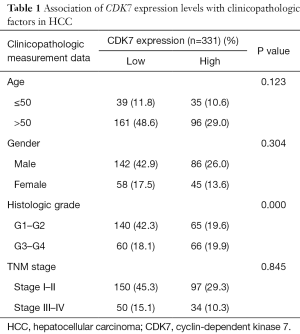
Full table
CDK7 expression level in HCC tissues and patients
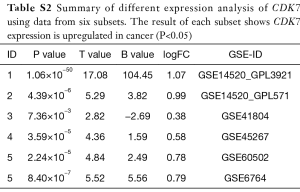
To investigate the expression level of CDK7 in HCC, we collected TCGA transcriptome data from the cBio Cancer Genomics Portal (www.cbioportal.org), including 375 liver-tumor and 50 non-tumor samples. The violin plot showed that CDK7 expression in the liver tumor was higher than in the non-tumor tissues (t-test =6.04, P=2.27×10−8, Figure 1C). Additionally, we examined the HCC transcriptome data in the GEO database to validate whether CDK7 was highly expressed in HCC tumor tissue. As a result, 650 samples (containing 341 tumors and 309 control tissues) were collected from five studies (GSE14520, GSE41804, GSE45267, GSE60502, and GSE6764). In each of the six subsets, CDK7 expression (probe code: 211297_s_at) was higher in the tumor tissues than in the para-carcinoma tissues (P<5×10−2, Table S2). Moreover, the result of the meta-analysis further confirmed that the expression level of CDK7 in liver cancer was higher than in the para-carcinoma tissues (Z score =−6.20, P=5.64×10−10, Figure 1D). Forest plots based on the five GEO datasets for comparing the expression of CDK7 in tumor and para-carcinoma tissues illustrated that these results were stable and without heterogeneity (SMD =1.32, 95% CI =1.02–1.62, P=7×10−2). Collectively, a high expression of CDK7 may play a crucial role in cancer progression and serve as a potential biomarker for cancer prognosis.
The influence of CDK7 downregulation on HCC cell lines
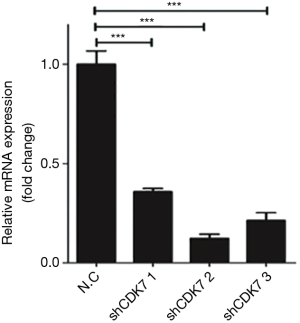
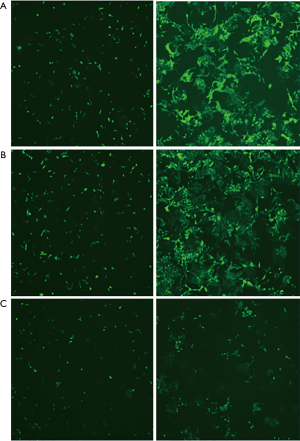
Two representative HCC cell lines (SMMC7721 and Huh7) were selected from a panel of eight HCC cell lines (MHHC97L, SMMC7721, HepG2, Hep3B, BEL-7402, Huh7, SK-hep and LM3) to determine the biological effect of CDK7 on HCC cells. A high expression of CDK7 was found in the SMMC7721 cell lines, while a low CDK7 expression was found in the Huh7 cell lines (Figure 1E). Subsequently, we evaluated the silencing capabilities of different CDK7 shRNAs and determined that CDK7-shRNA-2 decreased the expression level of CDK7 most conspicuously (Figure S2). CDK7 knockdown reduced the proliferation rate of the SMMC7721 and Huh7 cells as shown in Figure 1F,G and Figure S3. Both the SMMC-7721 cells and Huh7 cells, transfected with the CDK7 shRNA, showed significantly low growth capabilities during the 48-hour cell culturing period compared to the scrambled shRNA. Moreover, we found that the reduction of the proliferation capacity in the CDK7 shRNA-transfected Huh7 cells (P=2.33×10−3) was remarkably significant compared to that of the CDK7 shRNA-transfected SMMC-7721 cells (P=8.31×10−3), which might be because CDK7 is necessary for cell proliferation.
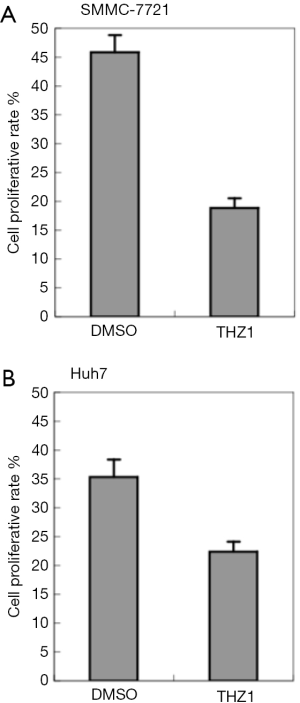
The effects of THZ1 on the HCC cell lines
THZ1 is a nanomolar intracellular CDK7 inhibitor that contains a cysteine-reactive acrylamide moiety and is reported to have a broad-based ability to target against the proliferation function of common cancer cell lines (9,23). SMMC-7721 and Huh7 cells were treated to THZ1 to assess the proliferation rate of the liver cancer cells exposed to the CDK7 inhibitor (Figure 2). The result showed THZ1 significantly inhibited the proliferation capacity of the SMMC-7721 (P=6.47×10−4) and Huh7 cells (P=2.52×10−2), suggesting that THZ1 might be a potent medicine for HCC treatment.
Conclusions
Based on the above analysis, we speculate that CDK7 may be a potential target for HCC treatment. It is logical to imply that THZ1, a potent irreversible covalent CDK7 inhibitor, is effective to treat HCC. THZ1 may lead to a considerable reduction in the phosphorylation status of the C-terminal domain of POLR2A, which partly explains its broad-based anti-proliferative activities and the possible mechanism of disturbing the global stability of mRNA. Furthermore, even with a small dose (50 nM), THZ1 causes the progressive loss of the critical apoptosis regulator RUNX1 and other genes regulated by RUNX1, including TAL1 and GATA, which play an important role in leukemia progression (9). The inhibition of CDK7 by THZ1 may inhibit the proliferation and progression of cancer cells by arresting the cell cycle at G2 phase, damaging the mitochondrion genes, and inhibiting the RTK/phosphatidylinositol 3 kinase/mitogen-activated protein kinase axis (10). Combined with what was discussed above, THZ1 has the potential to prevent HCC progression and spread as a next generation, ATP-site and allosteric irreversible covalent CDK7 inhibitor to repress HCC proliferation and induce HCC cell apoptosis.
In this study, we completed a systemic analysis of the relationship between CDK7 and the prognosis of multiple tumors, in particular, HCC. Extensively, the stable over-expression of CDK7 was observed in HCC, and it correlated to several clinicopathological features. Moreover, we confirmed that either the downregulation of CDK7 expression or treating HCC cells with its inhibitor THZ1 inhibited HCC cell proliferation. The potential role and mechanism of CDK7 were carefully analyzed and evaluated, suggesting a potential therapeutic target for THZ1.
Full table
Full table
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of Fujian Province (Grant No. #2014J01284 to C Zeng), the Education Department of Jiangxi Province (Grant No. #20151BAB215027, #20151BBG70255, and #2017ACB21005 to D Xu), and the National Natural Science Foundation of China (Grant No. #81760504 to F Fu).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.04.15). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- World Cancer Report 2014. World Health Organization, 2014. Chapter 5.6.
- Torre LA, Bray F, Siegel RL, et al. Global Cancer Statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development 2013;140:3079-93. [Crossref] [PubMed]
- Garriga J, Graña X. Cellular control of gene expression by T-type cyclin/CDK9 complexes. Gene 2004;337:15-23. [Crossref] [PubMed]
- Devos M, Mommaerts E, Migeot V, et al. Fission Yeast Cdk7 Controls Gene Expression through both Its CAK and C-Terminal Domain Kinase Activities. Mol Cell Biol 2015;35:1480-90. [Crossref] [PubMed]
- Wang Q, Li M, Zhang X, et al. Upregulation of CDK7 in gastric cancer cell promotes tumor cell proliferation and predicts poor prognosis. Exp Mol Pathol 2016;100:514-21. [Crossref] [PubMed]
- Christensen CL, Kwiatkowski N, Abraham BJ, et al. Targeting Transcriptional Addictions in Small Cell Lung Cancer with a Covalent CDK7 Inhibitor. Cancer Cell 2014;26:909-22. [Crossref] [PubMed]
- Chipumuro E, Marco E, Christensen CL, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 2014;159:1126-39. [Crossref] [PubMed]
- Kwiatkowski N, Zhang T, Rahl PB, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014;511:616-20. [Crossref] [PubMed]
- Greenall SA, Lim YC, Mitchell CB, et al. Cyclin-dependent kinase 7 is a therapeutic target in high-grade glioma. Oncogenesis 2017;6:e336 [Crossref] [PubMed]
- Zhang Z, Peng H, Wang X, et al. Preclinical efficacy and molecular mechanism of targeting CDK7-dependent transcriptional addiction in ovarian cancer. Mol Cancer Ther 2017;16:1739-50. [Crossref] [PubMed]
- Patel H, Abduljabbar R, Lai CF, et al. Expression of CDK7, Cyclin H, and MAT1 Is Elevated in Breast Cancer and Is Prognostic in Estrogen Receptor-Positive Breast Cancer. Clin Cancer Res 2016;22:5929-38. [Crossref] [PubMed]
- Naseh G, Mohammadifard M, Mohammadifard M. Upregulation of cyclin-dependent kinase 7 and matrix metalloproteinase-14 expression contribute to metastatic properties of gastric cancer. IUBMB Life 2016;799-805. [Crossref] [PubMed]
- Li B, Chonghaile TN, Fan Y, et al. Therapeutic rationale to target highly expressed CDK7 conferring poor outcomes in triple-negative breast cancer. Cancer Res 2017;77:3834-45. [Crossref] [PubMed]
- Zhang J, Yang X, Wang Y, et al. Low expression of CyclinH and cyclin-dependent kinase 7 can decrease the proliferation of human esophageal squamous cell carcinoma. Dig Dis Sci. 2013;58:2028-37. [Crossref] [PubMed]
- Davis S, Meltzer PS. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007;23:1846-7. [Crossref] [PubMed]
- Smyth GK. Limma: Linear Models for Microarray Data. In: Gentleman R, Carey V, Dudoit S, et al. editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer, 2005:397-420.
- Marot G, Foulley JL, Mayer CD, et al. Moderated effect size and P-value combinations for microarray meta-analyses. Bioinformatics 2009;25:2692-9. [Crossref] [PubMed]
- Khatri P, Roedder S, Kimura N, et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J Exp Med 2013;210:2205-21. [Crossref] [PubMed]
- Wang J, Coombes KR, Highsmith WE, et al. Differences in gene expression between B-cell chronic lymphocytic leukemia and normal B cells: A meta-analysis of three microarray studies. Bioinformatics 2004;20:3166-78. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Electronic address: wheeler@bcm.edu; Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327-1341.e23. [Crossref] [PubMed]
- Hoshida Y, Nijman SMB, Kobayashi M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res 2009;69:7385-92. [Crossref] [PubMed]
- Nilson KA, Guo J, Turek ME, et al. THZ1 Reveals Roles for Cdk7 in Co-transcriptional Capping and Pausing. Mol Cell 2015;59:576-87. [Crossref] [PubMed]


