
Epigenetic diagnostic biomarkers for non-small cell lung cancer: present and future perspectives
Introduction
Lung cancer is the most common cancer in China, accounting for an estimated 17.1% of all cancer diagnoses in 2015, and is the leading cause of cancer-related death in the country (1). Non-small cell lung cancer (NSCLC) accounts for about 85% of lung cancers and mainly presents as three subtypes: squamous cell carcinoma (SCC), adenocarcinoma (AD), and large cell carcinoma (LCC) (2). In 2015, the World Health Organization called for the classification of AD and SCC to include immunohistochemical staining to improve the accuracy of microscopic diagnoses (3,4). Most patients are asymptomatic at the early stages of lung cancer and diagnosis is often delayed until the diseased is at an advanced stage with tumor metastasis, leading to a poor prognosis (5). There are few effective early diagnostic markers for NSCLC, mainly due to its lack of symptoms and heterogeneity of clinical manifestations and pathologic features (6). Therefore, there is an urgent need to identify novel biomarkers that will help to increase the 5-year overall survival rate of NSCLC patients, which is currently less than 15% (7).
Accumulating evidence indicates that genetic and epigenetic changes regulate the development of cancer (8). Epigenetics refers to heritable changes in gene expression that are not due to alterations in the DNA coding sequence (9), such as DNA methylation, histone/nucleosome methylation and acetylation, and direct and indirect regulation by non-coding RNAs (ncRNAs). Interestingly, abnormal epigenetic regulation of gene expression is frequently associated with the development of many cancers, including NSCLC (10). Research on epigenetic modulators may therefore identify novel potential diagnostic and prognostic biomarkers for this disease.
Here, we review recent work on the potential clinical application of two classes of epigenetic regulators; DNA methylation and ncRNAs, both of which play a role in NSCLC tumorigenesis and may be a source of novel early diagnostic markers and therapeutic targets for NSCLC (Figure 1).
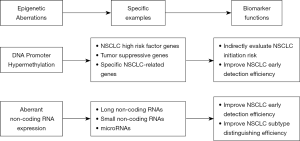
DNA methylation
Nuclear transcription factors regulate gene expression by binding to unique sequences in the gene promoter. The activity of transcription factors can be modulated by addition of methyl groups to specific nucleotides by DNA methyltransferases. This mainly occurs at sites enriched in cytosine and guanine dinucleotides (CpG, 5'-C-phosphate-G-3'), which can exist in an unmethylated state or be methylated at cytosine carbon-5. CpGs are scattered throughout the human genome but are often enriched in gene promoters as CpG islands, where they inhibit binding of transcription factors and consequently suppress gene transcription (11). Aberrant methylation of CpGs has been shown to contribute to the initiation and progression of cancer, suggesting that DNA methylation biomarkers could be useful in cancer diagnosis (12). In NSCLC, most DNA methylation occurs as gene-specific hypermethylation and genomic hypomethylation (13). Genes with potential clinical application in NSCLC mainly fall into three groups: NSCLC risk factor-related genes, universal tumor suppressor genes, and NSCLC-specific genes.
NSCLC risk factor-related genes
A number of factors carry a high risk for the development of NSCLC, including aging, cigarette smoking, and chronic inflammation, all of which have been shown to have epigenetic effects (14). Recent studies have identified specific methylated genes as potential NSCLC biomarkers, five of which (AHRR, F2RL3, 2q37.1, 6p21.33, and 12q14.1) have been shown to contain six CpG hypomethylation sites (14). The genes were identified by analysis of the peripheral blood of NSCLC patients, and their hypomethylation could be attributable to smoking. This finding suggests that modification of these genes may increase the risk of NSCLC and could thus be developed as predictive biomarkers (15).
Tumor suppressor genes
Inactivation of several well-known tumor suppressor genes through promoter methylation is an early event in the carcinogenic process of many cancers, including NSCLC (16). In one study, sputum samples collected from patients with SCC up to 3 years before clinical diagnosis showed evidence of aberrant methylation of the p16INK4a and/or O6-methylguanine-DNA methyltransferase gene (MGMT) promoters in 100% of samples (17). In another study, alterations (including methylation, mutation, or deletion) of the p16INK4a gene were observed in about 60% of NSCLC patients (18). Notably, methylation of p16INK4a is seen in all SCC patients and has been associated with smoking in AD patients (19). Considering the stability of the modification detected in samples and the accuracy with which it can be detected, DNA methylation of tumor suppressor genes holds great promise as a biomarker for the early diagnosis of NSCLC.
NSCLC-specific genes
In terms of feasibility for use as clinical biomarkers, targeted identification of specific methylated genes is a much preferable approach than genome-wide screening of methylation profiles. The methylation level of numerous genes plays a role in the initiation and progression of lung cancer, and can be easily detected by analysis of circulating tumor DNA (ctDNA) collected from the peripheral blood. ctDNAs are double-stranded DNA fragments ranging in length from 120 to 200 nucleotides (20). They are mostly released from apoptotic or necrotic tumor cells (21), with some originating from circulating tumor cells or released exosomes (22). Since methylated ctDNA is stable and can be collected in a convenient and simple manner, it has potential as an early diagnostic biomarker (23). For instance, analysis of peripheral blood-derived plasma showed aberrant methylation of the DCLK1 promoter in 32 of 65 (49.2%) NSCLC patients and only 8 of 95 (8.4%) of healthy controls (24). In another study, analysis of 330 plasma specimens from three independent case-control studies found that aberrant methylation of the SHOX2 and PTGER4 gene promoters could readily distinguish lung cancer patients from malignancy-free control subjects (25). Analysis of ctDNA from lung cancer patients has detected abnormal methylation levels in numerous genes, including short stature homeobox 2 (SHOX2), Ras association domain family 1 isoform (RASSF1A), retinoic acid receptor beta2 (RARB2), MGMT, and death-associated protein kinase 1 (DAPK) (26).
ncRNAs
Approximately 75% of the human genome is transcribed into RNA, but only a small proportion of that is translated into proteins (27). However, a large number of the remaining ncRNAs are actively involved in regulating cellular behavior through their interaction with RNA-binding proteins and regulation of target gene transcription. Importantly, many ncRNAs regulate the expression of oncogenes and tumor suppressor genes (28). Since ncRNAs can be readily identified and quantified by RT-PCR analysis of peripheral blood cell extracts, there is great interest in their potential clinical application in tumor diagnosis (29) After secreted into circulation, ncRNAs are extracted from blood samples of cancer patients and healthy controls by centrifugation. Consequently, the levels of ncRNAs are capable of being quantified and analyzed in duplicate by quantitative RT-PCR (30).
As a class, ncRNAs include housekeeping (transfer and ribosomal) RNAs and regulatory ncRNAs. The latter group can be further classified as long ncRNAs (lncRNAs >200 nucleotides) and short ncRNAs (<200 nucleotides) (31), which includes microRNAs (miRNAs), small interfering RNAs (siRNAs), small nucleolar RNAs (snoRNAs), PIWI-interacting RNAs (piRNAs), and small nuclear RNAs (snRNAs) (32).
Long ncRNAs
Epigenetic reprogramming of cancer cells causes a significant change in the profiles of lncRNAs with functions associated with tumor invasiveness and metastatic potential. Current work suggests that a number of lncRNAs could be used as diagnostic biomarkers for NSCLC.
HOX transcript antisense intergenic RNA (HOTAIR)
HOTAIR was the first lncRNA found to be involved in regulating tumorigenesis, and it has been shown to be dysregulated in several cancers, such as gastric cancer, breast cancer and NSCLC (33). HOTAIR is located within the homeobox C (HOXC) gene cluster on chromosome 12 and is co-expressed with HOXC (34). HOTAIR acts by binding to the polycomb repressive complex 2 (PRC2) and indirectly initiating trimethylation of H3 lysine 27 of the HOXD locus. In turn, this suppresses cell differentiation, enhances cell invasion and migration, and maintains cell stemness (35). HOTAIR-induced increases in cell invasiveness can be attributed to increased protein levels of the matrix metalloproteinases MMP2 and MMP9, which induce degradation of the extracellular matrix. Thus, tumor cells expressing high HOTAIR levels exhibit a high risk for metastasis, which correlates with poor prognosis and poor overall survival rates (36). Notably, HOTAIR levels are markedly higher in plasma and tumor tissues from NSCLC patients compared with normal subjects (37). In one study, HOTAIR levels could distinguish between NSCLC patients and healthy controls with relatively high sensitivity (76.2%) and specificity (71.9%) (34), confirming the potential utility of HOTAIR as a potential diagnostic biomarker.
Metastasis-associated lung ad transcript 1 (MALAT-1)
MALAT-1, which is located on chromosome 11q13 and is also known as nuclear-enriched abundant transcript 2 (NEAT2), is upregulated in several types of cancer including NSCLC (38). MALAT-1 is 8.7 kb in length and shows high species conservation (39). It regulates the expression of several genes related to cell motility, such as chaperonin containing TCP1 subunit 4 (CCT4), collagen triple helix repeat containing 1 (CTHRC1), hyaluronan mediated motility receptor (HMMR), and Regulator of differentiation 1 (ROD1), which indirectly enhances cell invasiveness. Thus, overexpression of MALAT-1 is significantly associated with the development of metastases (40). Exosome-derived MALAT-1 is significantly upregulated in sera from NSCLC patients compared with healthy controls (41). Interestingly, MALAT-1 levels can discriminate between SCC patients and AD patients and their respective controls with sensitivities of 63% and 48% (42), making it a promising epigenetic biomarker for the early diagnosis of NSCLC.
Colon cancer-associated transcript 2 (CCAT2)
Overexpression of this lncRNAs, which is transcribed from the 8q24 genomic region, is associated with the cell proliferation and invasiveness of various types of cancer (43). In a NSCLC cell line, expression of c-Myc and the miRNAs miR-17-5p and miR-20a were transcriptionally upregulated via CCAT2 and transcription factor 7 like 2 (TCF7L2), which leads to increased tumor growth (44). Moreover, CCAT2 is significantly overexpressed in NSCLC tissue compared with normal tissue (45,46), and its knockdown in NSCLC cell lines inhibited their proliferation and invasion in vivo and in vitro (45,47). Therefore, CCAT2 may be a promising biomarker for metastatic NSCLC.
NSCLC-specific lncRNAs
In addition to the lncRNAs described in the preceding sections, which regulate the behavior of various kinds of tumors, several lncRNAs have been discovered that are specifically dysregulated in NSCLC. For instance, the lncRNAs inactive X specific transcripts (XIST) and hypoxia inducible factor 1 alpha subunit antisense RNA 1 (HIF1A-AS1) (48) are upregulated in serum samples from NSCLC patients compared with controls. Moreover, aberrant expression of several lncRNAs is observed in different subtypes of NSCLC. Li et al. analyzed tissue microarrays from 181 patients with early-stage AD and found that LINC00313 is overexpressed only in patients with T2 and N1 stage AD (49). Zhang et al. found that LINC01133 is upregulated in samples of SCC, but not of AD (50), whereas Zhao et al. identified 72 aberrantly expressed lncRNAs in both SCC and AD patients (51). Since different NSCLC subtypes are associated with different pathological features and clinical treatment options, identification of lncRNAs capable of distinguishing between the NSCLC subtypes would represent a major advance in NSCLC diagnosis.
MicroRNAs
miRNAs are single-stranded ncRNAs, approximately 20 nucleotides in length, that show high species conservation (31). Since miRNAs are involved in virtually every aspect of cell physiology, it is not surprising that they are also important epigenetic regulators of the development and therapeutic response of NSCLC (52).
Recent studies have identified changes in the levels of several miRNAs in body fluids (e.g., serum, plasma) from cancer patients, highlighting the potential utility of this class of ncRNAs as diagnostic, prognostic, and/or predictive therapeutic response markers (53). Advances in microarray analytical techniques have led to the identification of multiple miRNAs with significant roles in the development of NSCLC. In one meta-analysis of 28 publications that analyzed 2,121 NSCLC patients and 1,582 healthy individuals, blood-derived miRNA levels could diagnose early disease with high accuracy (54). In another study of circulating miRNAs, 39 and 18 miRNAs were discovered to be upregulated and downregulated, respectively, in lung cancer patients compared with healthy controls, and were strongly associated with early or metastatic NSCLC (30). Assessment of panels of miRNA, rather than single miRNAs, will undoubtedly improve the specificity and sensitivity of NSCLC diagnosis (53).
Conclusions
Over the past several years, a broad spectrum of diagnostic biomarkers for NSCLC has been investigated. Here, we reviewed studies on two classes of potential epigenetic biomarkers. DNA promoter hypermethylation is an early event in NSCLC development, and primarily affects genes associated with a high risk of NSCLC, well-known tumor suppressor genes such as p16INK4a, and NSCLC-specific genes such as DCLK1. Most of these epigenetic changes can be detected by analyzing ctDNA in the peripheral circulation. In addition, technological advances in RNA sequencing and microarray analysis have identified several lncRNAs with potential utility for NSCLC diagnosis. Some of these, such as HOTAIR, MALAT-1, and CCAT2, are specifically associated with early-stage and/or metastatic disease, whereas others, such as LINC00313, are specifically overexpressed in a particular NSCLC subtype.
Notably, most epigenetic biomarkers can be readily detected through minimally or non-invasive approaches, which is an important feature for clinical applications. Moreover, combination of these epigenetic biomarkers and other early detection methods could have considerably higher diagnostic value than either method alone. From the data presented here, it seems reasonable to expect increasing focus on discovering epigenetic biomarkers for the diagnosis of NSCLC. Considering the rapid progress of technologies such as cloud computing and analysis of massive datasets, it should be feasible to construct an epigenetics database that includes tumors of many origins. Such a database would not only have clinical utility but also would help to advance our understanding of the initiation, progression, and regulation of cancer.
Acknowledgments
The work has not been published previously (except in the form of an abstract or as part of a published lecture or academic thesis or as an electronic preprint).
Funding: The study is supported by the Zhejiang Provincial Natural Science Foundation of China under Grant No. LY15H160034, China.
Footnote
Conflicts of Interest: TAll authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.05.03). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Devesa SS, Bray F, Vizcaino AP, et al. International lung cancer trends by histologic type: male:female differences diminishing and adenocarcinoma rates rising. Int J Cancer 2005;117:294-9. [Crossref] [PubMed]
- Inamura K. Lung Cancer: Understanding Its Molecular Pathology and the 2015 WHO Classification. Front Oncol 2017;7:193. [Crossref] [PubMed]
- Travis WD, Brambilla E, Burke AP, et al. Introduction to The 2015 World Health Organization Classification of Tumors of the Lung, Pleura, Thymus, and Heart. J Thorac Oncol 2015;10:1240-2. [Crossref] [PubMed]
- Ettinger DS, Akerley W, Borghaei H, et al. Non-small cell lung cancer, version 2.2013. J Natl Compr Canc Netw 2013;11:645-53; quiz 53. [Crossref] [PubMed]
- Hong QY, Wu GM, Qian GS, et al. Prevention and management of lung cancer in China. Cancer 2015;121:3080-8. [Crossref] [PubMed]
- Li A, Wei ZJ, Ding H, et al. Docetaxel versus docetaxel plus cisplatin for non-small-cell lung cancer: a meta-analysis of randomized clinical trials. Oncotarget 2017;8:57365-78. [PubMed]
- Geutjes EJ, Bajpe PK, Bernards R. Targeting the epigenome for treatment of cancer. Oncogene 2012;31:3827-44. [Crossref] [PubMed]
- Holliday R. The inheritance of epigenetic defects. Science 1987;238:163-70. [Crossref] [PubMed]
- Kanwal R, Gupta S. Epigenetics and cancer. J Appl Physiol (1985) 2010;109:598-605. [PubMed]
- Edwards JR, Yarychkivska O, Boulard M, et al. DNA methylation and DNA methyltransferases. Epigenetics Chromatin 2017;10:23. [Crossref] [PubMed]
- Lin SH, Wang J, Saintigny P, et al. Genes suppressed by DNA methylation in non-small cell lung cancer reveal the epigenetics of epithelial-mesenchymal transition. BMC Genomics 2014;15:1079. [Crossref] [PubMed]
- Hattori N, Ushijima T. Compendium of aberrant DNA methylation and histone modifications in cancer. Biochem Biophys Res Commun 2014;455:3-9. [Crossref] [PubMed]
- Georgiadis P, Hebels DG, Valavanis I, et al. Omics for prediction of environmental health effects: Blood leukocyte-based cross-omic profiling reliably predicts diseases associated with tobacco smoking. Sci Rep 2016;6:20544. [Crossref] [PubMed]
- Baglietto L, Ponzi E, Haycock P, et al. DNA methylation changes measured in pre-diagnostic peripheral blood samples are associated with smoking and lung cancer risk. Int J Cancer 2017;140:50-61. [Crossref] [PubMed]
- Tsou JA, Hagen JA, Carpenter CL, et al. DNA methylation analysis: a powerful new tool for lung cancer diagnosis. Oncogene 2002;21:5450-61. [Crossref] [PubMed]
- Palmisano WA, Divine KK, Saccomanno G, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res 2000;60:5954-8. [PubMed]
- Toyooka S, Mitsudomi T, Soh J, et al. Molecular oncology of lung cancer. Gen Thorac Cardiovasc Surg 2011;59:527-37. [Crossref] [PubMed]
- Toyooka S, Toyooka KO, Maruyama R, et al. DNA methylation profiles of lung tumors. Mol Cancer Ther 2001;1:61-7. [PubMed]
- Balgkouranidou I, Chimonidou M, Milaki G, et al. SOX17 promoter methylation in plasma circulating tumor DNA of patients with non-small cell lung cancer. Clin Chem Lab Med 2016;54:1385-93. [Crossref] [PubMed]
- Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Research 2001;61:1659-65. [PubMed]
- Shaw JA, Brown J, Coombes RC, et al. Circulating tumor cells and plasma DNA analysis in patients with indeterminate early or metastatic breast cancer. Biomark Med 2011;5:87-91. [Crossref] [PubMed]
- Wei SH, Balch C, Paik HH, et al. Prognostic DNA methylation biomarkers in ovarian cancer. Clin Cancer Res 2006;12:2788-94. [Crossref] [PubMed]
- Powrozek T, Krawczyk P, Nicos M, et al. Methylation of the DCLK1 promoter region in circulating free DNA and its prognostic value in lung cancer patients. Clin Transl Oncol 2016;18:398-404. [Crossref] [PubMed]
- Weiss G, Schlegel A, Kottwitz D, et al. Validation of the SHOX2/PTGER4 DNA Methylation Marker Panel for Plasma-Based Discrimination between Patients with Malignant and Nonmalignant Lung Disease. J Thorac Oncol 2017;12:77-84. [Crossref] [PubMed]
- Lu Y, Li S, Zhu S, et al. Methylated DNA/RNA in Body Fluids as Biomarkers for Lung Cancer. Biol Proced Online 2017;19:2. [Crossref] [PubMed]
- Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature 2012;489:101-8. [Crossref] [PubMed]
- Hauptman N, Glavac D. Long non-coding RNA in cancer. Int J Mol Sci 2013;14:4655-69. [Crossref] [PubMed]
- Ogata-Kawata H, Izumiya M, Kurioka D, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One 2014;9:e92921 [Crossref] [PubMed]
- Zhao C, Lu F, Chen H, et al. Clinical significance of circulating miRNA detection in lung cancer. Med Oncol 2016;33:41. [Crossref] [PubMed]
- Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the 'genome complexity' conundrum. Genes Dev 2007;21:11-42. [Crossref] [PubMed]
- Inamura K. Major Tumor Suppressor and Oncogenic Non-Coding RNAs: Clinical Relevance in Lung Cancer. Cells 2017;6: [Crossref] [PubMed]
- Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med 2015;12:1-9. [PubMed]
- Li N, Wang Y, Liu X, et al. Identification of Circulating Long Noncoding RNA HOTAIR as a Novel Biomarker for Diagnosis and Monitoring of Non-Small Cell Lung Cancer. Technol Cancer Res Treat 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Zhang J, Zhang P, Wang L, et al. Long non-coding RNA HOTAIR in carcinogenesis and metastasis. Acta Biochim Biophys Sin (Shanghai) 2014;46:1-5. [Crossref] [PubMed]
- Liu XH, Liu ZL, Sun M, et al. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer 2013;13:464. [Crossref] [PubMed]
- Liu MY, Li XQ, Gao TH, et al. Elevated HOTAIR expression associated with cisplatin resistance in non-small cell lung cancer patients. J Thorac Dis 2016;8:3314-22. [Crossref] [PubMed]
- Wu Y, Huang C, Meng XM, et al. Long Noncoding RNA MALAT1: Insights into its Biogenesis and Implications in Human Disease. Current Pharmaceutical Design 2015;21:5017-28. [Crossref] [PubMed]
- Tripathi V, Shen Z, Chakraborty A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet 2013;9:e1003368 [Crossref] [PubMed]
- Schmidt LH, Spieker T, Koschmieder S, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol 2011;6:1984-92. [Crossref] [PubMed]
- Zhang R, Xia Y, Wang Z, et al. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun 2017;490:406-14. [Crossref] [PubMed]
- Weber DG, Johnen G, Casjens S, et al. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res Notes 2013;6:518. [Crossref] [PubMed]
- Wang CY, Hua L, Yao KH, et al. Long non-coding RNA CCAT2 is up-regulated in gastric cancer and associated with poor prognosis. Int J Clin Exp Pathol 2015;8:779-85. [PubMed]
- Ling H, Spizzo R, Atlasi Y, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res 2013;23:1446-61. [Crossref] [PubMed]
- Zhao Z, Wang J, Wang S, et al. LncRNA CCAT2 promotes tumorigenesis by over-expressed Pokemon in non-small cell lung cancer. Biomed Pharmacother 2017;87:692-7. [Crossref] [PubMed]
- Katayama H, Tamai K, Shibuya R, et al. Long non-coding RNA HOTAIR promotes cell migration by upregulating insulin growth factor-binding protein 2 in renal cell carcinoma. Sci Rep 2017;7:12016. [Crossref] [PubMed]
- Qiu M, Xu Y, Yang X, et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol 2014;35:5375. [Crossref] [PubMed]
- Tantai J, Hu D, Yang Y, et al. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Pathol 2015;8:7887-95. [PubMed]
- Li M, Qiu M, Xu Y, et al. Differentially expressed protein-coding genes and long noncoding RNA in early-stage lung cancer. Tumour Biol 2015;36:9969-78. [Crossref] [PubMed]
- Zhang J, Zhu N, Chen X. A novel long noncoding RNA LINC01133 is upregulated in lung squamous cell cancer and predicts survival. Tumour Biol 2015;36:7465-71. [Crossref] [PubMed]
- Zhao W, Luo J, Jiao S. Comprehensive characterization of cancer subtype associated long non-coding RNAs and their clinical implications. Sci Rep 2014;4:6591. [Crossref] [PubMed]
- Inamura K, Ishikawa Y. MicroRNA In Lung Cancer: Novel Biomarkers and Potential Tools for Treatment. J Clin Med. 2016;5:36. [Crossref] [PubMed]
- Armand-Labit V, Pradines A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol Concepts 2017;8:61-81. [Crossref] [PubMed]
- Wang H, Wu S, Zhao L, et al. Clinical use of microRNAs as potential non-invasive biomarkers for detecting non-small cell lung cancer: a meta-analysis. Respirology 2015;20:56-65. [Crossref] [PubMed]


