
Circular RNAs and competing endogenous RNA (ceRNA) networks
What is an exonic circRNA?
Exonic circular RNAs (circRNAs) are covalently closed, single-stranded molecules produced by backsplicing of pre-mRNA transcripts (Figure 1) (1-3). Backsplicing is accomplished by the same splicesome machinery that generates mRNAs and exonic circRNAs carry 3',5'-phosphodiester bonds at their backsplice junctions. However, during circRNA synthesis, splicing does not follow the canonical 5'-3' order. Like linear mRNAs, exonic circRNAs are transported to the cytoplasm. Consistent with their circular nature, circRNAs lack the terminal structures [5' caps and poly(A) tails] associated with their linear counterparts and are resistant to exonucleases like RNAse R. Possibly because of their resistance to exonuclease digestion, circRNAs are generally more stable in vivo than linear transcripts.
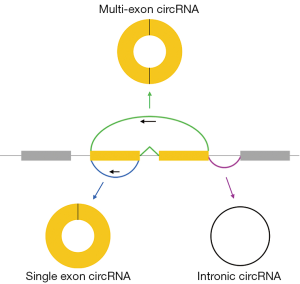
Intronic circular RNAs also exist. These are also formed by the normal spliceosome machinery but are biochemically distinct: they contain a 2'-5' linkage at their backsplice junctions. Intronic circRNAs remain nuclear and accordingly are proposed to have distinct functions from exonic circRNAs. This Perspective will focus specifically on exonic circRNAs.
Recent advances in genomic analyses have enabled systemic study of exonic circRNAs
circRNAs were first identified at least by 1991 (4) and in subsequent years several other circRNAs were fortuitously identified. However, major advances in the field occurred only recently upon application of novel and clever innovations to genome-wide analyses of RNA. In regard to sample prep, key technology advances include the use of ribosome depletion as an alternative to poly(A) purification for enrichment of mRNA and the use of RNAse R treatment to enrich for circular RNAs. Despite these enrichment steps, circRNAs can be difficult to identify/map and additional procedures are necessary to improve sensitivity. First, due to their low abundance, longer reads (100 bp or more) and increased depth of sequencing are essential to obtain sufficient sequence around the backsplice junctions. Second, sequencing samples before and after RNAse R treatment can help distinguish linear mRNA from circRNAs. Finally, novel computational pipelines take advantage of the non-canonical 3'-5' orientation of circRNA splicing. Combining these approaches, multiple labs have now characterized circRNAs across multiple cells types and species, and within various developmental and disease states. One very recent example of this type of analysis is described by Chen and colleagues who compared circRNA in fetal and adult muscle cells (5).
Several websites have been developed to catalog data collected from genomic analyses conducted across multiple labs including circBase (www.circbase.org), circ2Traits (http://gyanxet-beta.com/circdb/), circInteractome (https://circinteractome.nia.nih.gov/), circRNAdb (http://reprod.njmu.edu.cn/circrnadb), and PlantcircBase (http://ibi.zju.edu.cn/plantcircbase/).
General features of exonic circRNAs
circRNAs have been identified in plants, fungi, and in all metazoans examined, including C. elegans, Drosophila melanogaster, mice, and humans. In human fibroblasts, at least 10% of all transcribed genes generate circRNA isoforms (6). Across all cell types, it is likely that 50% of all genes have a circRNA isoform. Therefore, circRNAs are not oddities observed only at rare genes.
Several analyses suggest that in human cells, exonic circRNAs are present at about 1% of the levels of poly(A)RNA (3). These low levels at first suggested that circRNAs were unlikely to be functionally significant and that they existed only as a consequence of “errors” in the splicing process. However, more recent findings have provided evidence to suggest that circRNAs are functional. First, dozens of loci have been identified where circRNAs are more abundant than the linear isoforms (7). Second, many circRNAs are expressed in a cell type-, tissue-, developmental-, aging-, or disease-specific manner that does not always correlate with changes in the expression of the linear isoform (3). Third, conservation of circRNAs across species has been demonstrated (3). Fourth, and perhaps most interestingly, specific mechanisms for regulating expression of circRNA have been identified at some loci (8,9), demonstrating that at least in regards to these genes, evolution has acted to control how much circRNA is available in the cell.
Technically, it is now established that circRNA levels can be regulated by siRNA. This is important because it has allowed investigators to target specific circRNAs to directly test their role in normal cell function. Table 1 lists just a few examples of the many recent studies supporting functionality for specific circRNAs.
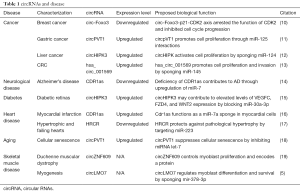
Full table
How can circRNAs affect cell function?
CircRNAs and peptide synthesis
About 40% of circRNAs carry AUG start codons that could in principle initiate translation of short peptides. In addition, investigators have demonstrated that engineered circular RNAs introduced into the cell can interact with ribosomes to generate protein (3). Moreover, at least one naturally occurring circRNA has been shown to direct peptide synthesis (19). However, in general, circRNAs are not associated with ribosomes and for now, peptide synthesis is not considered a likely role for most circRNAs. Instead, analysis of circRNA function has focused on their ability to interact with other RNAs to influence gene expression.
CircRNAs and competing endogenous RNA (ceRNA) networks
A profound breakthrough of recent genomic analyses has been the discovery that a significant proportion of the mammalian transcriptome does not correspond to annotated exons of protein-coding genes. Instead much of the transcriptional machinery is used to generate transcripts from pseudo-genes, long non-coding RNAs, and exonic circRNAs that do not encode peptides. Recent theoretical and experimental studies suggest that all these non-coding RNAs act to regulate each other and to regulate levels of coding transcripts by interacting with a limited pool of microRNAs (miRNAs) (Figure 2) (20,21).
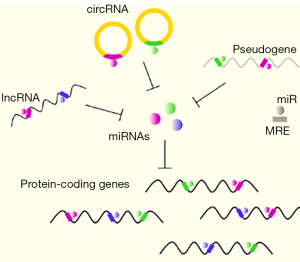
miRNAs are 22-nucleotide long species, processed from longer precursor mRNAs, that bind to 5–7 bases of complementary sequences on target RNAs to destabilize the target RNA and/or to reduce translation frequency (22,23). Thus miRNAs efficiently downregulate gene function. There are now thousands of miRNAs identified in mammalian cells. Target genes can have multiple miRNA binding sites (called MREs for miRNA response elements) and it is estimated that miRNAs thereby can regulate a large proportion of the transcriptome. In fact, miRNAs have been implicated in important developmental pathways, stress responses, and in numerous diseases including cancer.
circRNAs (like lncRNAs or pseudogenes) also carry MREs and thus have the ability to modulate miRNA activity by binding miRNA molecules and thereby reducing their availability to bind to and downregulate protein coding transcripts. This activity is often referred to as a “molecular sponge effect” or “sponging”. Because miRNAs invariably target multiple coding RNAs, a molecular sponge will affect expression of multiple genes. Thus circRNAs, along with their lincRNA and pseudogene accomplices, could form a large-scale regulatory network across the transcriptome.
Analysis of several model systems provides experimental support for the hypothesis that competition for miRNA regulates important biological functions. Many circRNAs described in Table 1 are proposed to work via titration of miRNAs. For example, Wei et al. indicate that circLMO7 regulates myoblast differentiation and survival by sponging mir-378-3p (5).
Despite this experimental support, there remain reasons for skepticism regarding the ability of many circRNAs to function as miRNA molecular sponges (24,25). Denzler et al. performed a very careful analysis of the ability of molecular sponge molecules to interfere with miR-122 targets and saw that sponging was observed only after the addition of >105 MRE sites per cell (24). These researchers suggest that MRE frequency per cell is already so high (partly because MREs are only 5–7 nucleotides and therefore present on many molecules) that expression of any single endogenous competitor would have to be extraordinarily high to be an effective sponge. As an alternative mechanism, it is interesting to consider that molecular sponges might work by regulating availability of RNA binding proteins (RBPs). RBPs might be more specific, in that they have fewer targets per cell than miRNAs, and therefore may be more susceptible to competition.
Summary
Advances in technologies for RNA deep sequencing and the development of specialized pipelines are allowing researchers to better characterize circRNA expression patterns. Some initial experiments support the idea that these molecules are more than random splicing errors and can have important biological functions. Currently much research is focused on circRNAs being part of a ceRNA network but continued work is required to determine just how important this process is to normal cell functioning.
Acknowledgments
Funding: This work was supported by the National Institutes of Health Division of Intramural Research (ZIA HD001804).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.05.12). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Greene J, Baird AM, Brady L, et al. Circular RNAs: biogenesis, function, and role in human diseases. Front Mol Biosci 2017;4:38. [Crossref] [PubMed]
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016;17:205-11. [Crossref] [PubMed]
- Jeck WR, Sharpless N. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Nigro JM, Cho K, Fearon E, et al. Scrambled exons. Cell 1991;64:607-13. [Crossref] [PubMed]
- Wei X, Li H, Yang J, et al. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis 2017;8:e3153 [Crossref] [PubMed]
- Jeck WR, Sorrentino J, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Salzman J, Chen R, Olsen M, et al. Cell-type specific features of circular RNA expression. PloS Genet 2013;9:e1003777 [Crossref] [PubMed]
- Ashwal-Fluss R, Meyer M, Pamudurti N, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55-66. [Crossref] [PubMed]
- Conn SJ, Pillman K, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015;160:1125-34. [Crossref] [PubMed]
- Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016;44:2846-58. [Crossref] [PubMed]
- Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett 2017;388:208-19. [Crossref] [PubMed]
- Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016;7:11215. [Crossref] [PubMed]
- Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 2016;7:26680-91. [PubMed]
- Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD). Front Genet 2013;4:307. [PubMed]
- Shan K, Liu C, Liu B, et al. Circular Noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation 2017;136:1629-42. [Crossref] [PubMed]
- Geng HH, Li R, Su Y, et al. The Circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on Its Target Genes Expression. PLoS One 2016;11:e0151753 [Crossref] [PubMed]
- Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016;37:2602-11. [Crossref] [PubMed]
- Panda AC, Grammatikakis I, Kim K, et al. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res 2017;45:4021-35. [Crossref] [PubMed]
- Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 2017;66:22-37.e9. [Crossref] [PubMed]
- Johnsson P, Ackley A, Vidarsdottir L, et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol 2013;20:440-6. [Crossref] [PubMed]
- Salmena L, Polseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell 2011;146:353-8. [Crossref] [PubMed]
- Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol 2010;17:1169-74. [Crossref] [PubMed]
- Bartel DP, Chen C. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 2004;5:396-400. [Crossref] [PubMed]
- Denzler R, Agarwal V, Stefano J, et al. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766-76. [Crossref] [PubMed]
- Denzler R, McGeary S, Title A, et al. Impact of MicroRNA levels, target-site complementarity, and cooperativity on competing endogenous RNA-regulated gene expression. Mol Cell 2016;64:565-79. [Crossref] [PubMed]


