
Exposing cancer with CRISPR-Cas9: from genetic identification to clinical therapy
Introduction
The system of clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9), first discovered in bacteria and archaea as an adaptive immune response to foreign genetic materials, has been extensively utilized as a precise genome editing tool for basic researches and therapeutic purposes in cancer (1,2). Cancer involves a single or multiple gene mutation(s), complex chromosomal alterations, translocations, losses and gains (3). Because the CRISPR-Cas9 system can recognize specific genomic loci via single guide RNAs (sgRNAs) and edit the loci effectively via the Cas9 protein, the accuracy and versatility of CRISPR-Cas9 system have been applied in biomedical research and to bring new opportunities to cancer research and treatment (4). Furthermore, the CRISPR-Cas9 system has the promising potential to accelerate progress in cancer research, whether by conducting functional genomics/epigenomics, modelling the genesis and progression of cancer in vitro and in vivo, targeting non-coding RNA, screening for novel therapeutic targets, or generating targeted cancer therapies and immunotherapies (1,5-7).
Depending on the delivery of viral or non-viral vectors, the CRISPR-Cas9 system can mediate precise genetic corrections or disruptions in in vitro and in vivo environments, and the CRISPR-Cas9 technology shows promise for improving outcomes for gene therapy (4,8-11). The CRISPR-Cas9 system has triggered enormous interest in therapeutic applications for cancers and exhibited the therapeutic potential in virally driven cancers (12,13).
CRISPR/Cas9 libraries have been constructed and applied for functional genomic screening and drug target discovery (14-16). CRISPR-based gene activation (CRISPRa) is highly useful in screening for gain of functions, and CRISPR-based gene inhibition (CRISPRi) is a more powerful tool than RNA interference (RNAi) in screening for loss of functions. Selection using CRISPR libraries can identify cancer survival-essential genes, which can be promising candidates for molecularly targeted drugs (14).
Cancer genomes commonly harbor multiple genetic aberrations which are the main factors that drive genesis and development of cancer, and the CRISPR-Cas9 system has been utilized to edit cancer-causing gene mutations and deletions and to engineer immune cells, such as chimeric antigen receptor T (CAR T) cells, for cancer immunotherapy (17). This review focuses on the application of CRISPR-Cas9 system for cancer research and therapy, including oncogene identification, genetic screen, druggable targets, cell or animal models, gene and cell therapy, and clinical trials. In addition, some concerns about translational CRISPR-Cas9 therapeutics are also discussed for cancer therapy.
The CRISPR-Cas9 system and oncogene identification
The CRISPR-Cas9 system has been utilized to validate or explore the function of cancer-associated genes via CRISPRi or CRISPRa. Wang et al. directly pool-mutagenized the livers of Cre-inducible CRISPR-Cas9 mice using adeno-associated viruses (AAVs) expressing a sgRNA library targeting putative tumor suppressor genes significantly mutated in human cancers. By molecular inversion probe sequencing of the sgRNA target sites to chart the mutational landscape of cancers, the authors revealed the functional consequence of multiple variants (for example, B2m and Kansl1) in driving liver tumorigenesis in immunocompetent mice (18). A member of solute carrier family 25 (SLC25A29), knocked out by CRISPR/Cas9 both in vitro and in vivo, was aberrantly elevated in cancer for modulating metabolic status to facilitate increased cancer progression (19). The CRISPR-Cas9 system was also employed to illustrate the relationship between p53 and metabolic stress-induced ferroptosis in cancer cells. The result suggested the p53-CDKN1A (p21) axis might help cancer cells cope with metabolic stress induced by cystine deprivation by delaying the onset of non-apoptotic cell death (20). The driver genes (KRAS, CDKN2A, SMAD4, and TP53) of pancreatic ductal adenocarcinoma were edited by the CRISPR-Cas9 system to demonstrate non-genetic acquisition of Wnt niche independence during pancreas tumorigenesis (21). The CRISPR-Cas9 system was adopted to comprehensively investigate epigenetic mechanisms, and Henser-Brownhill et al. generated a focused lentiviral-sgRNA library targeting 450 epigenetic regulators with multiple sgRNAs in human cells. The lentiviral-sgRNA library could be used not only for both selection and high-content screens, but also for targeted investigation of selected proteins without requiring isolation of knockout clones. After using a variety of functional assays, it was demonstrated the lentiviral-sgRNA library was suitable for both in vitro and in vivo applications, representing a new resource to study epigenetic mechanisms in physiological and pathological conditions (22). CRISPR-Cas9 screen revealed a MYCN-amplified neuroblastoma dependency on EZH2, which repressed neuronal differentiation in neuroblastoma in a PRC2-dependent manner. The data suggested that MYCN upregulated EZH2, leading to inactivation of a tumor suppressor program in neuroblastoma, and supported testing EZH2 inhibitors in MYCN-amplified neuroblastoma (23). RNA N6-methyladenosine methyltransferase METTL3 promoted liver cancer progression through YTHDF2 dependent post-transcriptional silencing of SOCS2. Using CRISPR/dCas9-VP64 activation system, Chen et al. demonstrated that overexpression of METTL3 significantly promoted human hepatocellular carcinoma growth both in vitro and in vivo. The findings revealed a new dimension of epigenetic alteration in liver carcinogenesis (24). CRISPR-dCas9 epigenetic editing tool was employed for simultaneous de novo DNA methylation of genes that commonly methylated in cancer. The promoter methylation prevented cells from engaging senescence arrest. The data showed that the key driver of this phenotype was repression of CDKN2A transcript p16 where myoepithelial cells harbored cancer-like gene expression. This study demonstrated that hit-and-run epigenetic events could prevent senescence entry, which might facilitate tumor initiation (25). Using CRISPR/Cas9-mediated ACVRIB-knockout and knockdown using siRNA, Loomans et al. found that loss of ACVRIB leaded to increased aggressiveness of squamous cell carcinoma of the head and neck and esophagus through alterations in cell-cell and cell-matrix adhesion proteins (26). Levels of Krüppel-like factor 5 (KLF5) were increased in human pancreatic ductal adenocarcinomas samples and in pancreatic intraepithelial neoplasia of Ptf1a-CreERTM;LSL-KrasG12D mice. KLF5 disruption by small hairpin RNAs or CRISPR/Cas9 strategies increased expression of NDRG2 and reduced activation of STAT3 and reduced acinar-to-ductal metaplasia and pancreatic intraepithelial neoplasia formation in mice. Strategies to reduce KLF5 activity might reduce progression of acinar cells from acinar-to-ductal metaplasia to pancreatic intraepithelial neoplasia and pancreatic tumorigenesis (27). Comparative genomics revealed that loss of lunatic fringe promoted melanoma metastasis, and CRISPR/Cas9-mediated knockout of lunatic fringe dramatically enhanced the capability of weakly metastatic melanoma cells to metastasize in vivo, a phenotype that could be rescued with the lunatic fringe cDNA. Therefore, lunatic fringe expression played a functional role in regulating melanoma metastasis (28). To analyze a prostate cancer risk region located at 7p15.2 to gain insight into the mechanisms by which the noncoding region might affect gene regulation and contribute to prostate cancer risk, the CRISPR-Cas9 system was exploited to delete a 4-kb region encompassing several prostate cancer risk-associated SNPs. The data suggested that the risk element regulated the expression of HOXA13 and HOTTIP via a repressive loop (29). Through a combination of CRISPR-Cas9-based genetic screening and metabolomic analyses, Romero et al. showed that KRAS/KEAP1- or KRAS/NRF2-mutant lung adenocarcinoma was dependent on increased glutaminolysis. Therefore, lung cancer harboring KRAS/KEAP1- or KRAS/NRF2-mutant might be treated by glutaminase inhibition (30). Dnajb1-Prkaca gene fusion in mice was generated by the CRISPR/Cas9 system, and Dnajb1-Prkaca gene fusion in wild-type mice was sufficient to initiate formation of tumors that displayed many features of human fibrolamellar hepatocellular carcinoma. The data suggested that strategies to block Dnajb1-Prkaca might be developed as therapeutics for this form of liver cancer (31). To address the potentially important roles of long noncoding RNAs, a genome-scale CRISPR-Cas9 activation screen that targeted more than 10,000 long noncoding RNAs transcriptional start sites was capitalized to identify noncoding loci that influenced a phenotype of interest. Joung et al. found 11 lncRNA loci that, upon recruitment of an activator, mediated resistance to BRAF inhibitors in human melanoma cells. This screen and characterization approach provided a CRISPR toolkit with which to systematically explore the functions of noncoding loci and elucidate their diverse roles in life sciences (32).
The CRISPR-Cas9 system and genetic screen for cancer
The CRISPR/Cas9 system is a powerful gene editing tool for gene knockout studies and functional genomic screens, and has revolutionized gene editing both at single genes and in multiplexed loss-of-function screens (33,34). The Hart lab created a database of Pooled In vitro CRISPR Knockout Library Essentiality Screens (PICKLES), where end users could display and download raw or normalized essentiality profiles for more than 18,000 protein-coding genes across more than 50 cell lines. Researchers could check the relative fitness defect and tissue specificity of their genes of interest, generate and save figures locally. The database is available at http://pickles.hart-lab.org. It should be useful in identifying tumor-specific essential genes for the development of targeted therapies (35). A computational method, CERES correction was developed to estimate gene-dependency levels from CRISPR-Cas9 essentiality screens while accounting for the copy number-specific effect. The results suggested that CERES correction decreased false-positive results in copy number-amplified regions. In addition, cancer-type-specific vulnerabilities could be identified after CERES correction (34). Cancer genomes commonly harbor hundreds of genetic aberrations which can be drivers or passengers of carcinogenesis and create vulnerabilities for potential therapeutic exploitation. To identify genotype-dependent vulnerabilities, forward genetic screens in different genetic backgrounds have been conducted. Rauscher et al. devised MINGLE, a computational framework to integrate CRISPR/Cas9 screens originating from different libraries. Researchers applied this method to analyze data from 85 CRISPR/Cas9 screens in human cancer cells to explore more than 2.1 million gene-background relationships. New genotype-specific vulnerabilities of cancer cells, GANAB and PRKCSH were identified as new positive regulators of Wnt/β-catenin signaling. By clustering genes with similar genetic interaction profiles, diverse genetic screens could be integrated to systematically build informative maps of genetic interactions in cancer (36). CRISPRa system was adapted for use in vivo to assess whether modulating endogenous gene expression levels could cause functional outcomes in the native environment of the liver. The in vivo CRISPRa platform allowed for parallel and combinatorial genetic screens in live animals and this approach enabled screening for drivers and suppressors of cell replication and tumor initiation (37). Combinatorial CRISPR-Cas9 screens were performed on a set of 51 carbohydrate metabolism genes, and it was revealed key nodes controlling redox homeostasis along the KEAP-NRF2 signaling axis. Furthermore, targeting KEAP1 gene alleviated the deleterious effects of these knockouts on growth rates. Therefore, the combinatorial CRISPR-Cas9 screens could be used to improve elucidation of metabolic network alterations and guide precision targeting of metabolic vulnerabilities based on tumor genetics (38). By CRISPR/Cas9-based targeted knockout for high-throughput screening of gene function in neuroblastoma cell differentiation, Long et al. found that plant homeodomain finger-containing protein 20 (PHF20) collaborated with poly (ADP-ribose) polymerase 1 (PARP1) to promote stemness and aggressiveness of neuroblastoma cells through activation of sex determining region Y-box 2 (SOX2) and octamer-binding transcription factor 4 (OCT4). PHF20 might be a new target for prognosis and therapy of neuroblastoma (39). Genome-wide CRISPR/Cas9 screens were used to identify mechanisms of resistance to TTK protein kinase inhibitors in multiple triple-negative breast cancer cell lines. Thu et al. discovered that the member of the anaphase-promoting complex/cyclosome (APC/C) complex was significantly associated with the response of TTK protein kinase inhibitors in breast and lung adenocarcinoma cell line panels. APC/C gene expression signature represented a potential biomarker that could be evaluated in ongoing clinical trials of TTK protein kinase inhibitors (40). PBAF, PBRM1 and ARID2 were identified as the associated genes of tumor cell resistance to killing by cytotoxic T cells via a genome-scale CRISPR-Cas9 screen (41). A genome-wide CRISPR/Cas9 enhancer/suppressor screen was performed in EGFR-dependent lung cancer PC9 cells to identify mechanistic insights and alternative drug escape pathways. The data revealed that dysregulation of ufmylation and endoplasmic reticulum (ER) stress comprised a previously unrecognized drug tolerance pathway that engaged survival signaling, with potentially important therapeutic implications (42). A high-throughput CRISPR-Cas9-based saturated mutagenesis approach, which could generate comprehensive libraries of point mutations, was used to predict clinical drug resistance with improved accuracy. All clinically isolated BCR-ABL1 mutations were identified via this CRISPR-Cas9-mediated saturated mutagenesis screen. The scientists believed that the strategy could be broadly applied to a variety of oncogenes to predict patient mutations and resistance susceptibility of drugs (43). To identify essential genes for cancer immunotherapy, a genome-scale CRISPR-Cas9 library that consisted of around 123,000 sgRNAs was capitalized to perturb genes in human melanoma cells to mimic loss-of-function mutations involved in resistance to T-cell-based immunotherapies. Patel et al. discovered multiple loss-of-function mutations in APLNR, encoding the apelin receptor. The functional loss of APLNR reduced the efficacy of adoptive cell transfer and checkpoint blockade immunotherapies in animal models (44).
The CRISPR-Cas9 system and druggable targets for cancer therapy
Mutagenesis studies and CRISPR-Cas9 screening showed the enzymatic SET domain of MLL/SET methyltransferases was not necessary for acute myeloid leukemia cell survival but that a newly identified domain termed the “FLOS” (functional location on SETD1A) was indispensable. FLOS disruption inhibited DNA damage response genes and induced p53-dependent apoptosis. Furthermore, the FLOS domain acted as a cyclin-K-binding site which was required for chromosomal recruitment of cyclin K in S phase. Therefore, targeting SETD1A and cyclin K complexes might represent a therapeutic opportunity for acute myeloid leukemia (45). The diphosphoinositol pentakisphosphate kinases (PPIP5Ks) could be potential targets for tumor therapy by CRISPR/Cas9 knockout (46). A genome-scale CRISPR knockout (GeCKO v2) library containing 123,411 sgRNAs was utilized to discover loss-of-function mutations conferring sorafenib resistance upon hepatocellular carcinoma cells. SGOL1 served as a druggable target for hepatocellular carcinoma treated with sorafenib and an indicator of prognosis via in vitro and in vivo study (47). CRISPR/Cas9 system was used to validate the Maternal Embryonic Leucine Zipper Kinase (MELK) as a cancer drug target, the results suggested that MELK expression correlated with tumor mitotic activity but was not required for cancer growth and that was not necessary for the proliferation of basal-like breast cancer cells (48,49). The cell cycle kinase MASTL acted as a critical role in cell cycle progression and might be a therapeutic target in human breast cancer by the evaluation based on CRISPRi and CRISPRa technologies. Also, genetic ablation of MASTL exhibited a significant therapeutic effect in vivo. MASTL might have both therapeutic and prognostic value in human breast cancer (50). CD38 was a promising target for cancer therapy. CRISPR/Cas9-based knockout of CD38 in human adenocarcinoma A549 cells inhibited anchorage-independent cell growth, invasion and xenograft growth in vivo. Consequently, CD38 played a role in human lung tumorigenesis and that anti-CD38 treatment might exhibit therapeutic potential in lung cancer (51). CMTM6 was a ubiquitously expressed protein that bound the programmed death-1 (PD-1) ligand 1 (PD-L1) and maintained its cell surface expression. CMTM6 could be a potential therapeutic target to overcome immune evasion by tumour cells because CMTM6 depletion decreased PD-L1 without compromising cell surface expression of MHC class I using a genome-wide CRISPR-Cas9 screen (52). Protein tyrosine phosphatase N23 (PTPN23), a suppressor of cell motility and invasion, was hemizygously or homozygously lost in breast cancer patients. In PTPN23-depleted tumors, Zhang et al. detected hyperphosphorylation of the autophosphorylation site tyrosine in the SRC family kinase FYN. Double knockout of FYN and PTPN23 via CRISPR-Cas9 system attenuated tumor outgrowth from PTPN23 knockout Cal51 cells. Therefore, FYN should be a therapeutic target for breast cancer with heterozygous or homozygous loss of PTPN23 (53). The protein tyrosine phosphatase PTPN2 was discovered via in vivo CRISPR-Cas9 screen and was a new immunotherapy target in unanticipated pathways, which could reverse resistance to PD-L1 immunotherapy by enhancing interferon-γ-mediated effects on antigen presentation and growth suppression (54). In melanoma cells that were both resistant and addicted to inhibition of the serine/threonine-protein kinase BRAF, an unbiased CRISPR-Cas9 knockout screen was performed to functionally mine their genome for “addiction genes”, which might be relayed by an ERK2-dependent phenotype switch (55). To identify genes affecting the tumor xenograft growth of human mutant KRAS (KRASMUT) colorectal cancers, the CRISPR-Cas9 system was exploited to conduct a genome-wide loss-of-function screen in an isogenic pair of human colorectal cancer cell lines harboring mutant or wild-type KRAS. The chromatin remodeling protein INO80C was identified as a novel tumor suppressor and might be a novel targetable set of therapeutic targets for KRASMUT tumors (56). Kodama et al. combined genome-wide CRISPR-Cas9 library and shRNA library screen to seek new therapeutic targets for epithelial ovarian cancer. KPNB1, a novel oncogene, was highly expressed in patients with epithelial ovarian cancer and associated with recurrence and poor prognosis. Furthermore, the combination of paclitaxel and ivermectin, a KPNB1-dependent antitumor drug, produced a strong antitumor effect on epithelial ovarian cancer both in vitro and in vivo. The data identified a combinatorial therapy for epithelial ovarian cancer, in addition to a plethora of potential drug targets (57).
The CRISPR-Cas9 system and generation of cell or animal models for cancer researches
To validate the function of target gene(s), the CRISPR-Cas9 system was commonly employed to generate gene-knockout cells or animal models. FoxO3A−/− cancer cells were established using CRISPR/Cas9 genome editing system to reveal the novel function of the transcription factor FoxO3A in mitochondria of cancer cells (58). CRISPR-generated ID8 ovarian cancer cell models with single (Trp53−/−), double (Trp53−/−;Brca2−/−), (Trp53−/−;Brca1−/−), (Trp53−/−;Nf1−/−) and (Trp53−/−;Pten−/−), and triple (Trp53−/−;Brca2−/−;Pten−/−) suppressor gene deletions might represent a new and simple tool to investigate the biology of ovarian high grade serous carcinoma (59,60). The tumor suppressor gene adenomatous polyposis coli (APC) was mutated in most patients with colorectal cancers, resulting in constitutive Wnt activation. Novellasdemunt et al. engineer various APC-truncated isogenic lines using CRISPR/Cas9 technology to investigate the Wnt-activating mechanism of the APC mutation. β-catenin inhibitory domain (CID)-deleted APC truncation promoted β-catenin deubiquitination. USP7-deleted APC truncation suppressed Wnt activation by restoring β-catenin ubiquitination, and inhibited xenograft tumor growth. Therefore, USP7 was a tumor-specific Wnt activator for APC-mutated colorectal cancer by mediating β-catenin deubiquitination (61). Slc25a11∆/∆ immortalized mouse chromaffin knockout cells generated by CRISPR-Cas9 system was used to illustrate that the germline mutation in the mitochondrial 2-oxoglutarate/malate carrier SLC25A11 gene conferred a vulnerability to metastatic paragangliomas (62). The IMD-KO (Adm2−/−) mice were generated by the CRISPR/Cas9 system to study the mechanisms of intermedin in vascular remodeling and blood perfusion. In murine retinas, intermedin significantly promoted vascular lumen enlargement by reactivating the quiescent endothelial cells to proliferate, which contributed to the improved blood perfusion in both physiological (retinal) and pathological (tumor) angiogenic models (63). To elucidate the role of cell cycle-related kinase (CCRK) on immunosuppression in hepatocellular carcinoma, liver-specific CCRK-inducible transgenic mice were generated by CRISPR/Cas9-mediated CCRK depletion. The results suggested that tumorous CCRK depletion could diminish myeloid-derived suppressor cell immunosuppression and enhance immune-checkpoint blockade efficacy by upregulating PD-L1 expression and increasing intratumorous CD8+ T cells (64). In addition, the CRISPR-Cas9 system was applied to delete key DNA repair genes in human colon organoids to study the origin of mutational signatures in cancer (65).
The CRISPR-Cas9 system and cancer gene therapy in vivo
CRISPR-Cas9 technology has been applied for cancer gene therapy in animal models. He et al. constructed a folate-modified liposome to deliver CRISPR-Cas9 plasmid targeting DNA methyltransferase 1 (DNMT1) gene for ovarian cancer gene therapy. The liposome/CRISPR plasmid complex could express Cas9 protein, downregulate DNMT1 in tumor tissue and inhibit tumor growth in vivo. This DNMT1 gene-targeted and folate receptor-targeted CRISPR-Cas9 delivery system might be a potential therapeutic regimen for ovarian cancer (66-68). Cancer-derived exosomes efficiently deliver CRISPR-Cas9 plasmids expressing sgRNA targeting poly (ADP-ribose) polymerase-1 (PARP-1) to ovarian cancer tumors of SKOV-3 xenograft mice. CRISPR-Cas9-loaded exosomes inhibited expression of PARP-1, leading to the induction of apoptosis in ovarian cancer cells. Furthermore, the inhibition of PARP-1 increased the chemosensitivity to cisplatin, showing synergistic cytotoxicity. Therefore, CRISPR-Cas9-loaded exosomes might be very promising for genome editing therapeutics of cancer in the future (69). Optogenetic and CRISPR/Cas9 techniques were introduced into a recombinant adenovirus (Ad) vector to produce an illumination-dependent spatiotemporally controllable gene regulation system (Opt/Cas-Ad system). This Opt/Cas-Ad system regulated a tumor suppressor gene effectively in a xenograft tumor model. Furthermore, tumor treatment could be viewed by illuminating the tumor and be stopped simply by turning off the light. The Opt/Cas-Ad system might enhance both the safety and effectiveness of cancer gene therapy (70). sgRNA targeted mutant KRAS alleles was screened and validated in cancer cells. Intra-tumoral injection of lentivirus and AAV coding Cas9 and sgRNA inhibited tumor growth in immunodeficient mice. The results suggested that CRISPR-Cas9 system could be utilized to target the KRAS driver mutation of cancers in vitro and in vivo (71). A TAT peptide-modified Au nanoparticle was utilized to condense CRISPR-Cas9 plasmid coding sgRNA targeting Plk-1, a master regulator gene of mitosis, to suppress Plk-1 expression for cancer therapy. The TAT-Au-CRISPR plasmid complex was coated with cationic and neutral lipids to form a delivery system (LACP) with shell of lipids and core of TAT-Au-CRISPR plasmid complex. LACP displayed long-term stability in blood and long-term circulation capability in mice by intravenous injection. Also, LACP via intratumoral injection inhibited tumor growth effectively by tumor cell apoptosis. This multiple component assembly might provide a versatile method for CRISPR-Cas9 system delivery and targeted genome editing for therapeutics of cancer (72). Vascular endothelial growth factor A (VEGFA) was a promising therapeutic target for osteosarcoma. Osteosarcoma cell-specific aptamer (LC09)-functionalized PEG-PEI-Cholesterol lipopolymer (PPC) was synthesized to encapsulate CRISPR-Cas9 plasmids encoding VEGFA sgRNA and CRIPSR-Cas9 for prepare LC09-PPC/Cas9-VEGFA complexes. LC09-PPC/Cas9-VEGFA complexes facilitated selective delivery of CRISPR-Cas9 plasmid in both orthotopic osteosarcoma and lung metastasis, resulting in effective VEGFA disruption in tumor tissues, inhibited orthotopic osteosarcoma malignancy and lung metastasis. This novel in vivo tumor-targeted delivery system might be developed as a potential alternate for genome editing therapeutics of osteosarcoma (73).
The CRISPR-Cas9 system and cell therapy for cancer
Cas9 protein and sgRNA were co-delivered by arginine nanoparticles into macrophages to generate SIRP-α knockout macrophages. The SIRP-α knockout macrophages could not receive the “don’t eat me signal” signal from cancer cells and improve attack and elimination of cancer cells. This strategy has the potential for development of “weaponized” macrophages for cancer immunotherapy (74). Adoptive transfer of T cells genetically modified to express a cancer-specific T-cell receptor (TCR) has exhibited therapeutic potential for cancer. However, the endogenous TCRs might compete with transgenic TCR in the recipient cells, which affected the efficacy of cancer immunotherapy. Endogenous TCR-β was knocked out from the recipient cells using CRISPR-Cas9 system, and then the TCR-β T cells was transduced with a cancer-reactive receptor of choice to produce transgenic αβ and γδ TCRs. The TCR-plus-CRISPR-modified T cells were up to a thousand-fold more sensitive to cancer antigen than standard TCR-transduced T cells. Consequently, compared with standard TCR transfer, the γδ TCR-plus-CRISPR-modified T cells resulted in more efficient redirection of CD4+ and CD8+ T cells against a panel of established blood cancers and patient-derived B-cell acute lymphoblastic leukemia. The results suggested that TCR transfer combined with CRISPR-Cas9 genome editing could yield new, improved generations for cancer immunotherapies (75). The CAR T cells were edited by CRISPR-Cas9 system to generate universal CAR T cells and potent T cells that are resistant to exhaustion and inhibition. CRISPR-Cas9 system might promote the application of CAR T cells in cancer immunotherapy (17,76).
The CRISPR-Cas9 system and clinical trials for cancer therapy
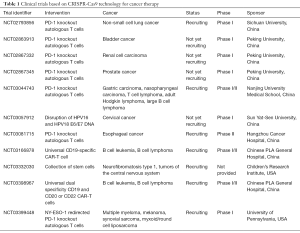
Since the scientists and oncologists from West China Hospital declared they would apply CRISPR-Cas9 technique to human subjects in clinical trials in 2016 (77), there have been 14 studies found in ClinicalTrials.gov database up to now, and the indications of 11 clinical trials among the 14 studies are cancer, including cervical cancer, multiple myeloma, melanoma, synovial sarcoma, myxoid/round cell liposarcoma, B cell leukemia, esophageal cancer, neurofibromatosis type 1, tumors of the central nervous system, bladder cancer, prostate cancer, renal cell carcinoma, non-small cell lung cancer, gastric carcinoma, nasopharyngeal carcinoma, T cell lymphoma, adult Hodgkin lymphoma, large B cell lymphoma (Table 1). In these clinical trials, CRISPR-Cas9 system was applied to edit the autologous T cells from patients and the edited T cells were infused back into patients. This strategy, or with modification, was adopted by almost all designers of clinical trials. The cancer therapy based on CRISPR-Cas9 system might provide a new approach for cases who had no choice after standard therapies (78,79).
Full table
Perspectives
The CRISPR-Cas9 technology is a powerful tool for cancer research and therapy. With various CRISPR-Cas9 libraries and databases, virtual or computational high-throughput screens can hit some essential genes for carcinogenesis, progression, metastasis and tolerance. Next, the functions of genes (candidates) are also identified in cell or animal models established by CRISPRa or CRISPRi, and then gene medicines or small molecule drugs are designed and evaluated for advanced cancer therapy. Although some new druggable targets have been discovered via the CRISPR-Cas9 system, most of them are at the preclinical stage. When delivering CRISPR-Cas9 plasmids, mRNA, Cas9 protein and sgRNAs for cancer gene therapy, it is concerned the delivery vectors with basic requirements of high efficiencies and good safety in vivo. Furthermore, almost all in vivo studies using non-viral vectors for cancer gene therapy did not addressed off-target effects of the CRISPR-Cas9 system. Therefore, advanced delivery system needs to be further developed for the CRISPR-Cas9 system, and the off-target effects should be explored for cancer gene therapy. Immune cells are edited for cancer immunotherapies by the CRISPR-Cas9 system, which is the most promising approach for the clinical based upon genome editing therapeutics. PD-1 knockout autologous T cell and CAR T cell therapies based on the CRISPR-Cas9 technique have been entered the clinic trials in China and USA, and the explored genome editing therapeutics are being accepted by scientists and the public. The efficacy, off-targets and ethics are obstacles for the CRISPR-Cas9 therapeutics, but they will be solved with the development of sciences and technologies. To conclude, it is believed that there are translational candidates, based on the CRISPR-Cas9 technique, from bench to bed for cancer therapy.
Acknowledgments
Funding: This work was supported by the National Natural and Scientific Foundation of China (NO. 81602699), the Science & Technology Department of Sichuan Province (NO. 2018GZ0311), the Health and Family Planning Commission of Sichuan Province (NO. 18PJ543), the National High Technology Research and Development Program of China (NO. 2015AA020309), and the China Postdoctoral Science Foundation Funded Project (NO. 2015M570791).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.06.16). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Aquino-Jarquin G. Emerging role of CRISPR/Cas9 technology for microRNAs editing in cancer research. Cancer Res 2017;77:6812-7. [Crossref] [PubMed]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014;157:1262-78. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Ratan ZA, Son YJ, Haidere MF, et al. CRISPR-Cas9: a promising genetic engineering approach in cancer research. Ther Adv Med Oncol 2018;10:1758834018755089 [Crossref] [PubMed]
- Moses C, Garcia-Bloj B, Harvey AR, et al. Hallmarks of cancer: The CRISPR generation. Eur J Cancer 2018;93:10-8. [Crossref] [PubMed]
- Buyel JF. Plants as sources of natural and recombinant anti-cancer agents. Biotechnol Adv 2018;36:506-20. [Crossref] [PubMed]
- Luo X, Qiu Y, Jiang Y, et al. Long non-coding RNA implicated in the invasion and metastasis of head and neck cancer: possible function and mechanisms. Mol Cancer 2018;17:14. [Crossref] [PubMed]
- Beitelshees M, Hill A, Rostami P, et al. Pressing diseases that represent promising targets for gene therapy. Discov Med 2017;24:313-22. [PubMed]
- He ZY, Men K, Qin Z, et al. Non-viral and viral delivery systems for CRISPR-Cas9 technology in the biomedical field. Sci China Life Sci 2017;60:458-67. [Crossref] [PubMed]
- Men K, Duan X, He Z, et al. CRISPR/Cas9-mediated correction of human genetic disease. Sci China Life Sci 2017;60:447-57. [Crossref] [PubMed]
- Li L, Song L, Liu X, et al. Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano 2017;11:95-111. [Crossref] [PubMed]
- Liu C, Zhang L, Liu H, et al. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release 2017;266:17-26. [Crossref] [PubMed]
- Jubair L, McMillan NAJ. The therapeutic potential of CRISPR/Cas9 systems in oncogene-addicted cancer types: Virally driven cancers as a model system. Mol Ther Nucleic Acids 2017;8:56-63. [Crossref] [PubMed]
- Kurata M, Yamamoto K, Moriarity BS, et al. CRISPR/Cas9 library screening for drug target discovery. J Hum Genet 2018;63:179-86. [Crossref] [PubMed]
- Poirier JT. CRISPR libraries and screening. Prog Mol Biol Transl Sci 2017;152:69-82. [Crossref] [PubMed]
- Sharifnia T, Hong AL, Painter CA, et al. Emerging opportunities for target discovery in rare cancers. Cell Chem Biol 2017;24:1075-91. [Crossref] [PubMed]
- Wu HY, Cao CY. The application of CRISPR-Cas9 genome editing tool in cancer immunotherapy. Brief Funct Genomics 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Wang G, Chow RD, Ye L, et al. Mapping a functional cancer genome atlas of tumor suppressors in mouse liver using AAV-CRISPR-mediated direct in vivo screening. Sci Adv 2018;4:eaao5508.
- Zhang H, Wang Q, Gu J, et al. Elevated mitochondrial SLC25A29 in cancer modulates metabolic status by increasing mitochondria-derived nitric oxide. Oncogene 2018;37:2545-58. [Crossref] [PubMed]
- Tarangelo A, Magtanong L, Bieging-Rolett KT, et al. p53 suppresses metabolic stress-induced ferroptosis in cancer cells. Cell Rep 2018;22:569-75. [Crossref] [PubMed]
- Seino T, Kawasaki S, Shimokawa M, et al. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell 2018;22:454-67.e6. [Crossref] [PubMed]
- Henser-Brownhill T, Monserrat J, Scaffidi P. Generation of an arrayed CRISPR-Cas9 library targeting epigenetic regulators: from high-content screens to in vivo assays. Epigenetics 2017;12:1065-75. [Crossref] [PubMed]
- Chen L, Alexe G, Dharia NV, et al. CRISPR-Cas9 screen reveals a MYCN-amplified neuroblastoma dependency on EZH2. J Clin Invest 2018;128:446-62. [Crossref] [PubMed]
- Chen M, Wei L, Law CT, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 2018;67:2254-70. [Crossref] [PubMed]
- Saunderson EA, Stepper P, Gomm JJ, et al. Hit-and-run epigenetic editing prevents senescence entry in primary breast cells from healthy donors. Nat Commun 2017;8:1450. [Crossref] [PubMed]
- Loomans HA, Arnold SA, Hebron K, et al. Loss of ACVRIB leads to increased squamous cell carcinoma aggressiveness through alterations in cell-cell and cell-matrix adhesion proteins. Am J Cancer Res 2017;7:2422-37. [PubMed]
- He P, Yang JW, Yang VW, et al. Krüppel-like factor 5, increased in pancreatic ductal adenocarcinoma, promotes proliferation, acinar to ductal metaplasia, pancreatic intraepithelial neoplasia, and tumor growth in mice. Gastroenterology 2018;154:1494-508.e13. [Crossref] [PubMed]
- Del Castillo Velasco-Herrera M, van der Weyden L, Nsengimana J, et al. Comparative genomics reveals that loss of lunatic fringe (LFNG) promotes melanoma metastasis. Mol Oncol 2018;12:239-55. [Crossref] [PubMed]
- Luo Z, Rhie SK, Lay FD, et al. A prostate cancer risk element functions as a repressive loop that regulates HOXA13. Cell Rep 2017;21:1411-7. [Crossref] [PubMed]
- Romero R, Sayin VI, Davidson SM, et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med 2017;23:1362-8. [PubMed]
- Engelholm LH, Riaz A, Serra D, et al. CRISPR/Cas9 engineering of adult mouse liver demonstrates that the Dnajb1-Prkaca gene fusion is sufficient to induce tumors resembling fibrolamellar hepatocellular carcinoma. Gastroenterology 2017;153:1662-73.e10. [Crossref] [PubMed]
- Joung J, Engreitz JM, Konermann S, et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 2017;548:343-6. [Crossref] [PubMed]
- Yuen G, Khan FJ, Gao S, et al. CRISPR/Cas9-mediated gene knockout is insensitive to target copy number but is dependent on guide RNA potency and Cas9/sgRNA threshold expression level. Nucleic Acids Res 2017;45:12039-53. [Crossref] [PubMed]
- Meyers RM, Bryan JG, McFarland JM, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet 2017;49:1779-84. [Crossref] [PubMed]
- Lenoir WF, Lim TL, Hart T. PICKLES: the database of pooled in-vitro CRISPR knockout library essentiality screens. Nucleic Acids Res 2018;46:D776-80. [Crossref] [PubMed]
- Rauscher B, Heigwer F, Henkel L, et al. Toward an integrated map of genetic interactions in cancer cells. Mol Syst Biol 2018;14:e7656 [Crossref] [PubMed]
- Wangensteen KJ, Wang YJ, Dou Z, et al. Combinatorial genetics in liver repopulation and carcinogenesis with a novel in vivo CRISPR activation platform. Hepatology 2017; [Epub ahead of print]. [PubMed]
- Zhao D, Badur MG, Luebeck J, et al. Combinatorial CRISPR-Cas9 metabolic screens reveal critical redox control points dependent on the KEAP1-NRF2 regulatory axis. Mol Cell 2018;69:699-708.e7. [Crossref] [PubMed]
- Long W, Zhao W, Ning B, et al. PHF20 collaborates with PARP1 to promote stemness and aggressiveness of neuroblastoma cells through activation of SOX2 and OCT4. J Mol Cell Biol 2018;10:147-60. [Crossref] [PubMed]
- Thu KL, Silvester J, Elliott MJ, et al. Disruption of the anaphase-promoting complex confers resistance to TTK inhibitors in triple-negative breast cancer. Proc Natl Acad Sci U S A 2018;115:E1570-7. [Crossref] [PubMed]
- Pan D, Kobayashi A, Jiang P, et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 2018;359:770-5. [Crossref] [PubMed]
- Terai H, Kitajima S, Potter DS, et al. ER stress signaling promotes the survival of cancer "persister cells" tolerant to EGFR tyrosine kinase inhibitors. Cancer Res 2018;78:1044-57. [Crossref] [PubMed]
- Ma L, Boucher JI, Paulsen J, et al. CRISPR-Cas9-mediated saturated mutagenesis screen predicts clinical drug resistance with improved accuracy. Proc Natl Acad Sci U S A 2017;114:11751-6. [Crossref] [PubMed]
- Patel SJ, Sanjana NE, Kishton RJ, et al. Identification of essential genes for cancer immunotherapy. Nature 2017;548:537-42. [Crossref] [PubMed]
- Hoshii T, Cifani P, Feng Z, et al. A non-catalytic function of SETD1A regulates cyclin K and the DNA damage responseS. Cell 2018;172:1007-21.e17. [Crossref] [PubMed]
- Gu C, Nguyen HN, Ganini D, et al. KO of 5-InsP7 kinase activity transforms the HCT116 colon cancer cell line into a hypermetabolic, growth-inhibited phenotype. Proc Natl Acad Sci U S A 2017;114:11968-73. [Crossref] [PubMed]
- Sun W, He B, Yang B, et al. Genome-wide CRISPR screen reveals SGOL1 as a druggable target of sorafenib-treated hepatocellular carcinoma. Lab Invest 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Giuliano CJ, Lin A, Smith JC, et al. MELK expression correlates with tumor mitotic activity but is not required for cancer growth. Elife 2018;7: [Crossref] [PubMed]
- Huang HT, Seo HS, Zhang T, et al. MELK is not necessary for the proliferation of basal-like breast cancer cells. Elife 2017;6: [Crossref] [PubMed]
- Álvarez-Fernández M, Sanz-Flores M, Sanz-Castillo B, et al. Therapeutic relevance of the PP2A-B55 inhibitory kinase MASTL/Greatwall in breast cancer. Cell Death Differ 2018;25:828-40. [Crossref] [PubMed]
- Bu X, Kato J, Hong JA, et al. CD38 knockout suppresses tumorigenesis in mice and clonogenic growth of human lung cancer cells. Carcinogenesis 2018;39:242-51. [Crossref] [PubMed]
- Burr ML, Sparbier CE, Chan YC, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 2017;549:101-5. [Crossref] [PubMed]
- Zhang S, Fan G, Hao Y, et al. Suppression of protein tyrosine phosphatase N23 predisposes to breast tumorigenesis via activation of FYN kinase. Genes Dev 2017;31:1939-57. [Crossref] [PubMed]
- Manguso RT, Pope HW, Zimmer MD, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 2017;547:413-8. [Crossref] [PubMed]
- Kong X, Kuilman T, Shahrabi A, et al. Cancer drug addiction is relayed by an ERK2-dependent phenotype switch. Nature 2017;550:270-4. [Crossref] [PubMed]
- Yau EH, Kummetha IR, Lichinchi G, et al. Genome-wide CRISPR screen for essential cell growth mediators in mutant KRAS colorectal cancers. Cancer Res 2017;77:6330-9. [Crossref] [PubMed]
- Kodama M, Kodama T, Newberg JY, et al. In vivo loss-of-function screens identify KPNB1 as a new druggable oncogene in epithelial ovarian cancer. Proc Natl Acad Sci U S A 2017;114:E7301-10. [Crossref] [PubMed]
- Celestini V, Tezil T, Russo L, et al. Uncoupling FoxO3A mitochondrial and nuclear functions in cancer cells undergoing metabolic stress and chemotherapy. Cell Death Dis 2018;9:231. [Crossref] [PubMed]
- Walton JB, Farquharson M, Mason S, et al. CRISPR/Cas9-derived models of ovarian high grade serous carcinoma targeting Brca1, Pten and Nf1, and correlation with platinum sensitivity. Sci Rep 2017;7:16827. [Crossref] [PubMed]
- Walton J, Blagih J, Ennis D, et al. CRISPR/Cas9-mediated Trp53 and Brca2 knockout to generate improved murine models of ovarian high-grade serous carcinoma. Cancer Res 2016;76:6118-29. [Crossref] [PubMed]
- Novellasdemunt L, Foglizzo V, Cuadrado L, et al. USP7 is a tumor-specific WNT activator for APC-mutated colorectal cancer by mediating beta-catenin deubiquitination. Cell Rep 2017;21:612-27. [Crossref] [PubMed]
- Buffet A, Morin A, Castro-Vega LJ, et al. Germline mutations in the mitochondrial 2-oxoglutarate/malate carrier SLC25A11 gene confer a predisposition to metastatic paragangliomas. Cancer Res 2018;78:1914-22. [Crossref] [PubMed]
- Wang LJ, Xiao F, Kong LM, et al. Intermedin enlarges the vascular lumen by inducing the quiescent endothelial cell proliferation. Arterioscler Thromb Vasc Biol 2018;38:398-413. [Crossref] [PubMed]
- Zhou J, Liu M, Sun H, et al. Hepatoma-intrinsic CCRK inhibition diminishes myeloid-derived suppressor cell immunosuppression and enhances immune-checkpoint blockade efficacy. Gut 2018;67:931-44. [Crossref] [PubMed]
- Drost J, van Boxtel R, Blokzijl F, et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 2017;358:234-8. [Crossref] [PubMed]
- He ZY, Wei XW, Luo M, et al. Folate-linked lipoplexes for short hairpin RNA targeting claudin-3 delivery in ovarian cancer xenografts. J Control Release 2013;172:679-89. [Crossref] [PubMed]
- He ZY, Zhang YG, Yang YH, et al. In vivo ovarian cancer gene therapy using CRISPR-Cas9. Hum Gene Ther 2018;29:223-33. [Crossref] [PubMed]
- He ZY, Deng F, Wei XW, et al. Ovarian cancer treatment with a tumor-targeting and gene expression-controllable lipoplex. Sci Rep 2016;6:23764. [Crossref] [PubMed]
- Kim SM, Yang Y, Oh SJ, et al. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J Control Release 2017;266:8-16. [Crossref] [PubMed]
- Takayama K, Mizuguchi H. Generation of optogenetically modified adenovirus vector for spatiotemporally controllable gene therapy. ACS Chem Biol 2018;13:449-54. [Crossref] [PubMed]
- Kim W, Lee S, Kim HS, et al. Targeting mutant KRAS with CRISPR-Cas9 controls tumor growth. Genome Res 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Wang P, Zhang L, Zheng W, et al. Thermo-triggered release of CRISPR-Cas9 system by lipid-encapsulated gold nanoparticles for tumor therapy. Angew Chem Int Ed Engl 2018;57:1491-6. [Crossref] [PubMed]
- Liang C, Li F, Wang L, et al. Tumor cell-targeted delivery of CRISPR/Cas9 by aptamer-functionalized lipopolymer for therapeutic genome editing of VEGFA in osteosarcoma. Biomaterials 2017;147:68-85. [Crossref] [PubMed]
- Ray M, Lee YW, Hardie J, et al. CRISPRed macrophages for cell-based cancer immunotherapy. Bioconjug Chem 2018;29:445-50. [Crossref] [PubMed]
- Legut M, Dolton G, Mian AA, et al. CRISPR-mediated TCR replacement generates superior anticancer transgenic T cells. Blood 2018;131:311-22. [Crossref] [PubMed]
- Ren J, Zhao Y. Advancing chimeric antigen receptor T cell therapy with CRISPR/Cas9. Protein Cell 2017;8:634-43. [Crossref] [PubMed]
- Cyranoski D. Chinese scientists to pioneer first human CRISPR trial. Nature 2016;535:476-7. [Crossref] [PubMed]
- Normile D. China sprints ahead in CRISPR therapy race. Science 2017;358:20-1. [Crossref] [PubMed]
- Baylis F, McLeod M. First-in-human phase 1 CRISPR gene editing cancer trials: Are we ready? Curr Gene Ther 2017;17:309-19. [PubMed]


