
ND4 mutations are more prevalent in patients with acute myeloid leukemia of M2 morphology
Introduction
Acute myeloid leukemia (AML) is a group of heterogeneous diseases with varying clinical characteristics and prognostic implications. Conventional genetic prognostic markers, including FLT3-ITD, NPM1, c-KIT, CEBPA, DNMT3A, and MLL-PTD, combined with cytogenetic abnormalities are considered to be the important prognostic factors of AML (1,2). In patients with a normal karyotype, specific and accurate predictors may play a key role in the evaluation of prognosis. These normal cytogenetic patients with isolated bi-allelic CEBPA (biCEBPA) or NPM1 mutation without FLT3-ITD are associated with a favorable prognosis, whereas FLT3-ITD or TP53 mutations are associated with a poor prognosis (3-6). DNA methyltransferase 3A (DNMT3A) gene mutations are associated with hyperleukocytosis at disease presentation, the elderly, and a poor prognosis (7). Patients less than 60 years and with DNMT3A, FLT3-ITD, and NPM1 mutations had a shorter event-free survival (EFS) (P=0.047). Further, patients with DNMT3A and NPM1 mutations had a significantly shorter overall survival (OS) compared to those with FLT3-ITD and NPM1 mutations (P=0.047) suggesting that the adverse impact of DNMT3A mutations is more pronounced than that of FLT3-ITD among patients with an NPM1 mutation (8).
Mitochondrial dysfunction has been found in cancer progression (9). Human mitochondrial DNA (mtDNA) is a 16-kb circle that contains genes encoding 13 electron transport chain proteins, 22 tRNAs, and 2 rRNAs (10). Disrupted electron transport chain function was due to mtDNA mutations, which involve mitochondrial genes encoding components of respiratory Complex I of the electron transport chain. ND4, one of the seven Complex I subunits encoded by mtDNA, is predicted to be important for proton translocation (9). Acquired deletions of mtDNA in the hematopoietic compartment have also been found to occur in association with some hematological diseases (10). Mutations in the mitochondrial NADH dehydrogenase subunit 4 (ND4) were described in three of 93 AML patients, but the importance of these mutations is not yet clear (11). Another study noted 29 of 452 patients (6.4%) had ND4 mutations predicted to affect translation, which implied acquired ND4 mutations in AML may have a favorable prognostic value. Patients with somatically acquired ND4 mutations had significantly longer relapse-free survival (RFS) and OS than ND4 wild-type patients, while germline ND4 mutations tended to have shorter survival (12). The clinical characteristics of ND4 gene mutations were not very clear and the prognostic influence in AML was not validated until now. We performed this study to systematically investigate the frequency and the prognostic relevance of new molecular markers and conventional gene mutations in 460 adult AML patients and divide these patients into appropriate prognostic groups using molecular markers.
Methods
Patients and treatment
From 2004 to 2016, bone marrow (BM) and peripheral blood (PB) samples were collected from 460 patients diagnosed with de novo AML according to the French-American-British (FAB) criteria admitted to the First Affiliated Hospital of Nanjing Medical University, Jiangsu Province Hospital. Two hundred and sixty patients were males and 200 were females, with a median age of 47 years (range, 18–86 years). Median follow-up time was 18 months (range, 1–144 months). The diagnosis of core binding factor (CBF) AML was based on cytogenetic findings of karyotype t(8;21) and inv[16] or detection of the fusion transcripts AML1-ETO and CBFβ-MYH11 by reverse transcription polymerase chain reaction (RT-PCR). This study was approved by the ethics board of the hospital.
Gene mutations and cytogenetic analysis
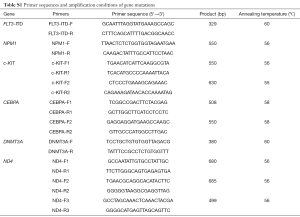
Genomic DNA was isolated according to standard protocol. For mutation analysis, the whole amplicon or hot spot of FLT3-ITD, NPM1, c-KIT, CEBPA, DNMT3A, and ND4 genes were amplified using standard PCR conditions (Table S1). Somatic or germline status of ND4 mutations was established by evaluating matched samples, with follow-up samples obtained when patients were in complete remission (CR). The mutational status of these genes was determined by Sanger sequencing. The BM samples of the patients were studied mostly by R-banding analysis, and chromosomal abnormalities were described according to the International System for Human Cytogenetic Nomenclature 2013 (ISCN2013).
Flow cytometric analysis
Cell surface antigens were analyzed by four-color flow cytometry. Anti-CD34, human leukocyte antigen (HLA)-DR, CD117, CD13, CD33, CD14, CD15, CD10, CD19, CD20, CD22, CD2, CD3, CD5, CD7, CD4, and CD8 were purchased from BD Pharmingen (San Diego, CA, USA). All cell surface antigens were detected following the manufacturer’s instructions and the data were analyzed using CELLQuest software (Becton-Dickinson).
Treatment protocols
For acute promyelocytic leukemia (APL) patients with PML-RARα fusion gene or t(15;17), all-trans retinoic acid and arsenic trioxide -based treatment was given for the induction and consolidation therapy. Other AML patients received standard first-line treatment. In the consolidation therapy group, patients were additionally treated with high-dose cytarabine-based chemotherapy (13).
Statistical analysis
Quantitative data were described in the form of mean ± standard deviation (SD). Statistical analysis was done by comparison between groups using one-way ANOVA regarding quantitative data and chi-square test regarding qualitative data while quantitative nonparametric data comparison was performed using Mann-Whitney U. Kaplan-Meier analysis was used to calculate the distribution of OS and RFS. All P values <0.05 (two-tailed) were considered statistically significant. The statistical analysis of data was done by using SPSS version 17.0.
Results
Frequencies of gene mutations
Among 460 AML patients, FLT3-ITD mutations were found in 48 (48/385, 12.5%), NPM1 in 67 (67/428, 15.7%), c-KIT in 7 (7/312, 2.2%), mono-allelic CEBPA (monoCEBPA) in 37 (37/287, 12.9%), bi-allelic CEBPA (biCEBPA) in 10 (10/287, 3.5%), DNMT3A in 20 (20/252, 7.9%), and ND4 in 10 (10/152, 6.6%).
Fifty-two patients (11.3%) were diagnosed with CBF AML. There were 6 patients with inv[16] or CBFβ-MYH11 and 46 patients with t(8;21) or AML1-ETO. FLT3-ITD (4/20, 20.0%) were frequently present in addition to the PML-RARα fusion in the M3 subtype, but there was no significant difference among different groups (P>0.05 for all comparisons). Conversely, c-KIT mutations were most commonly seen in CBF leukemia patients (6 of 37, 16.2%) and all the cases were of the M2 type. Patients with FLT3-ITD, NPM1, CEBPA, or DNMT3A mutations had no significant difference in the distribution of CBF, M3, or non-CBF and M3 patients (P>0.05) while patients with c-KIT mutations were more frequent in CBF patients than in non-CBF and M3 patients (P<0.001). DNMT3A mutations were found in 18.4% (7/38) and 10.7% (3/28) of M5 and M4 subtype, respectively. There was a significant difference of DNMT3A mutations in M5 but not in M4 (P=0.006 and 0.498, respectively). In patients with intermediate cytogenetic risk, there were six cases of M5, five cases of M2, two cases of M1, and two cases of M4 with DNMT3A mutations. There was no significant difference of distribution in M5 and M4 between patients with DNMT3A mutations and patients with intermediate cytogenetic risk and DNMT3A mutations (P=0.134 and 1.0, respectively).
Types of ND4 mutations and correlations with other molecular markers
The mutation types of ND4 were A131V, F149L, G242D, A404T, and Y409H. The most common amino-acid change of ND4 mutation was the A404T substitution, which was observed in four cases (4/10, 40.0%). The A131V mutation was observed in three patients, while F149L, G242D, and Y409H mutations were only detected in a single case. The germline mutations included were types of A131V, F149L, A404T, and Y409H in non-M3 patients, while one M3 patient had somatic mutation with type of G242D. These mutations were established by evaluating matched samples, with follow-up samples obtained when patients were in CR.
In the ND4 mutated patients, three patients had molecular abnormalities including monoCEBPA and biCEBPA mutations. One patient had the A131V mutation, while another one patient had the Y409H mutation, and both these patients had an additional monoCEBPA mutation. One patient with A404T mutation had a biCEBPA mutation, while the other mutated patients had no additional mutations.
Clinical characteristics of patients with gene mutations
In non-M3 patients, NPM1 (P<0.001) and DNMT3A (P=0.012) mutations were older than patients with the wild type, while patients with c-KIT mutations were younger (P=0.003) (Table 1). FLT3-ITD (P<0.001) and NPM1 (P=0.003) mutations were associated with high white blood cell (WBC) counts, while c-KIT (P<0.001) mutations were associated with low WBC counts. FLT3-ITD (P=0.005), NPM1 (P=0.001), and monoCEBPA (P=0.013) mutations were associated with a higher percentage of blasts in the BM (Table 1). There was no significant difference in any of the gene mutations according to gender, level of hemoglobin (HB) or platelet (PLT) counts (P>0.05 for all comparisons).
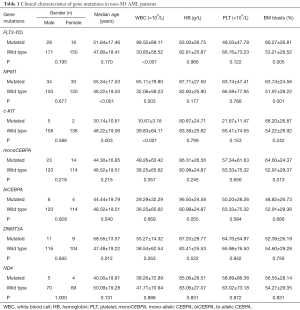
Full table
Of a total of ten patients with ND4 mutations, three had normal cytogenetics, three others had favorable cytogenetics including t(8;21) or t(15;17), one patient had a complex karyotype, another one patient had a non-defined karyotype, while no karyotype was found in the remaining two patients. Six patients were males and four were females. Patients with ND4 mutations were M1 (2/10, 20.0%), M2 (7/10, 70.0%), and M3 (1/10, 10.0%) types based on the FAB classification: three patients with A131V were M2; four patients with A404T were: three M2 and one M1; one patient with F149L was M2; one patient with G242D was M3, and one patient with Y409H was M1. ND4 mutations were prevalent in patients with AML of M2 morphology (P=0.001). Flow cytometric analysis of leukemic cells showed CD34 positive (7/10, 70.0%), CD117 positive (10/10, 100%), HLA-DR positive (9/10, 90.0%), CD13 positive (9/10, 90.0%), CD33 positive (9/10, 90.0%), CD15 positive (7/10, 70.0%), and T- or B-associated markers CD4 and/or CD7 and/or CD19 positive (5/10, 50.0%) (data not shown). The median age of ND4 mutated patients vs. ND4 wild-type patients in non-M3 patients was 40 vs. 50 years (P=0.131). In non-M3 patients, patients with ND4 mutations were associated with higher expression of CD19 (P=0.045). Compared with patients with ND4 wild-type, patients with ND4 mutations were not associated with FLT3-ITD, NPM1, c-KIT, CEBPA, and DNMT3A mutations in non-M3 patients (P>0.05).
Response to induction therapy and survival analysis
Rate of CR was 62.9%, and older patients (P=0.032), and males (P=0.003) were significantly associated with lower rates of CR in non-M3 patients. Relapse rate (RR) of non-M3 patients was 25.4% (P=0.001) and CBF patients was 24.1% (P=0.004), while it was 0 in M3 patients.
About 60% of patients (6/10) with ND4 mutations achieved CR, and of the 10 patients, 4 patients had a relapse. Compared with patients with ND4 wild-type, patients with ND4 mutations had no significant difference in CR rate or RRs (P>0.05).
In the OS and RFS analysis for patients with ND4 mutations and wild-type ND4, the median OS were 13 and 18 months, respectively, while the RFS median were 11 and 16 months, respectively. In non-M3 patients, patients with ND4 germline mutations may have unfavorable prognosis, but there was no statistical significance in OS and RFS between patients with germline ND4 mutations and wild-type patients (P=0.159 and 0.087, respectively) (Figure 1A,B).
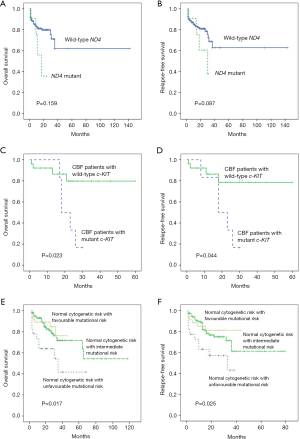
We then compared the OS and RFS of CBF AML patients according to c-KIT mutation status. There was a significant difference in OS and RFS between c-KIT-mutated and wild-type patients in Kaplan-Meier analysis (P=0.023 and 0.044, respectively) (Figure 1C,D). CBF AML patients with c-KIT mutations had poor prognosis compared with patients of wild-type c-KIT.
Among patients with normal cytogenetic AML, we classified patients into three categories: patients with NPM1 mutation or biCEBPA mutation without FLT3-ITD mutations; patients with FLT3-ITD mutations; patients with another molecular profile. Mutational status could further stratify AML patients with normal cytogenetic risk into three subgroups, which was a supplement of the cytogenetic prognosis risk (P=0.017 and 0.025 for OS and RFS, respectively) (Figure 1E,F). The favorable mutational risk demonstrated the longest survival and vice versa with unfavorable mutational risk.
In the univariate analysis, patients older than 60 years [HR for OS, 1.684 (1.097–2.585), P=0.017; HR for RFS, 1.638 (1.067–2.515), P=0.024], FLT3-ITD [HR for OS, 2.065 (1.210–3.524), P=0.008; HR for RFS, 2.013 (1.176–3.446), P=0.011], favorable molecular group [HR for OS, 0.641 (0.434–0.948), P=0.026; HR for RFS, 0.662 (0.446–0.983), P=0.041] and normal cytogenetic risk with favorable mutational risk [HR for OS, 0.528 (0.301–0.928), P=0.026; HR for RFS, 0.544 (0.313–0.945), P=0.0.031] were related to prognosis of non-M3 patients. There was no statistical significance in OS and RFS in multivariate analysis of any parameters (P>0.05 for all comparisons).
Discussion
About 50% of AML patients had no cytogenetic markers which are independent predictors for prognosis (14,15), genetic mutations may play an important role in the prognostic value in AML. Different molecular markers usually stratify AML patients into subtypes with distinctive prognosis and response to therapy (16). Recent descriptions of mutations in the mitochondrial ND4 gene in leukemia patients coupled with altered metabolic function to leukemogenesis (10). In this study, we attempted to study the value of mutation of ND4 and DNMT3A and other conventional factors in the prognosis of AML patients and the role of cytogenetic and molecular markers in the evaluation of prognosis.
Shen et al. (13) believed that there was subtype-restricted distribution in AML, such as c-KIT mutation, often as the second hit, which plays an important role in the pathogenesis of CBF leukemia. We found that patients with c-KIT mutations were more frequent in CBF patients than in non-CBF and M3 patients (P<0.001). Yan et al. (17) discovered DNMT3A mutations were 20.5% and 13.6% of M5 and M4 subtype, respectively. Our results showed that DNMT3A mutations were found in M5 but not in M4 (P=0.006 and 0.498, respectively), indicating that DNMT3A mutations are restricted to the monocytic lineage involvement in AML. Marková et al. (18) showed that occurrence of DNMT3A mutations was not associated with particular FAB subtypes in patients with intermediate-risk cytogenetics. Our results also showed that patients with intermediate-risk cytogenetics and with DNMT3A mutations had no significant difference in distribution of M5 and M4 subtypes.
ND4 is a part of respiratory Complex I, which leads to decreased Complex I activity and subsequently decreased NAD+ generation may also result in altered α-ketoglutarate production and epigenetic modulation (12). There were 11 predicted transmembrane domains of ND4 may be important for mitochondrial proton transport and four types (F149L, G242D, A404T and Y409H) of the mutations we detected were within these domains. ND4 mutations affect mitochondrial function, and the survival and/or proliferative capacities of leukemia cells may be changed (12). In this study, patients with ND4 mutations were considered as M1 (20.0%), M2 (70.0%), and M3 (10.0%) types based on FAB classification. ND4 mutations were prevalent in M2 patients, and this must be further validated. Damm et al. (12) observed that ND4 mutated patients tended to be younger (P=0.059), with a median age of 39 vs. 47 years for ND4 wild-type patients. Our results showed no significant difference in non-M3 patients. The non-M3 patients have germline mutation types of A131V, F149L, A404T, and Y409H, while one M3 patient had somatic mutation with type of G242D. This was in line with the conclusion of the study by Damm et al. (12). In non-M3 patients, ND4 mutations were associated with higher expression of CD19 (P=0.045). A close relationship was observed between the expression of CD19 and t(8;21) (19), and certain AML1-ETO-positive cases demonstrated characteristic immunological features (such as CD19 and CD34 expressions, and CD33 negativity) (20). Hence, we speculated that ND4 mutations were found more often in M1 and M2 patient types in this study, and were associated with the expression of CD19.
In the present study, RR of non-M3 patients was 25.4% (P=0.001) and CBF of patients was 24.1% (P=0.004), while it was 0 in M3 patients, indicating that M3 patients always had a favorable response to therapy. Patients with ND4 germline mutations may have unfavorable prognosis, but there was no statistical significance in OS and RFS between patients with germline ND4 mutations and wild-type patients (P=0.159 and 0.087, respectively; Figure 1A,B). Patients with somatic ND4 mutations had favorable prognosis than ND4 wild-type patients (P<0.05 for both comparisons), while no significant differences were found in patients with ND4 mutations and ND4 wild-type patients (12). Obviously, compared to patients with ND4 wild-type, patients with somatic ND4 mutations demonstrated favorable prognosis and patients with germline ND4 mutations tended to have poor prognosis.
Boissel et al. (21) found that c-KIT mutations were associated with a shorter EFS and RFS (P=0.002 and 0.003) in t(8;21) but not in inv[16] patients. We compared the OS and RFS of CBF AML patients according to c-KIT mutation status and found that there was a significant difference in OS and RFS between c-KIT-mutated and wild-type patients (P=0.023 and 0.044, respectively) (Figure 1C,D). In the present study, we tried to stratify patients with normal cytogenetics into different subgroups. We found that, according to the molecular prognostic factors, patients with normal cytogenetic risk could be divided into three groups in the OS and RFS analysis (P=0.017 and 0.025, respectively) (Figure 1E,F). Different mutational risks have distinctive prognosis in these groups, favorable mutational risk had the longest survival and unfavorable mutational risk had the shortest survival. The significance for prognosis could be observed, which in turn could help the physicians to treat patients in an individualized manner.
Conclusions
In summary, combining the cytogenetic risk and the mutational risk, an important role in the distinctive stratification of AML patients is possible. ND4 mutations were prevalent in M2 patients and were associated with higher expression of CD19. Whether patients with germline ND4 mutations have poorer prognosis still needs to be confirmed.
Full table
Acknowledgments
Funding: This work was sponsored by grants from the National Natural Science Foundation of China (No. 81400095, 81570134, 81570141, 81522001, 81400079, 81600096), Suzhou Industrial Technology Innovation Research Project (SYSD2017004), Jiangsu Provincial Health and Family Planning Commission Medical Research Project (H2017087), the Project of National Key Clinical Specialty, the National Science & Technology Pillar Program (2014BAI09B12), and National Science and Technology Major Project (2017ZX09304032).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.07.21). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Our study was approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (2018-SR-136) and because the medical records/biological specimens were obtained from previous clinical diagnosis, the exemption of informed consent was approved by the IRB.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Luskin MR, Lee JW, Fernandez HF, et al. Benefit of high-dose daunorubicin in AML induction extends across cytogenetic and molecular groups. Blood 2016;127:1551-8. [Crossref] [PubMed]
- Bochtler T, Fröhling S, Krämer A. Role of chromosomal aberrations in clonal diversity and progression of acute myeloid leukemia. Leukemia 2015;29:1243-52. [Crossref] [PubMed]
- Ahn JS, Kim JY, Kim HJ, et al. Normal karyotype acute myeloid leukemia patients with CEBPA double mutation have a favorable prognosis but no survival benefit from allogeneic stem cell transplant. Ann Hematol 2016;95:301-10. [Crossref] [PubMed]
- Alpermann T, Schnittger S, Eder C, et al. Molecular subtypes of NPM1 mutations have different clinical profiles, specific patterns of accompanying molecular mutations and varying outcomes in intermediate risk acute myeloid leukemia. Haematologica 2016;101:e55-8. [Crossref] [PubMed]
- Deol A, Sengsayadeth S, Ahn KW, et al. Does FLT3 mutation impact survival after hematopoietic stem cell transplantation for acute myeloid leukemia? A Center for International Blood and Marrow Transplant Research (CIBMTR) analysis. Cancer 2016;122:3005-14. [Crossref] [PubMed]
- Welch JS, Petti AA, Miller CA, et al. TP53 and Decitabine in Acute Myeloid Leukemia and Myelodysplastic Syndromes. N Engl J Med 2016;375:2023-36. [Crossref] [PubMed]
- Zare-Abdollahi D, Safari S, Movafagh A, et al. A mutational and expressional analysis of DNMT3A in acute myeloid leukemia cytogenetic subgroups. Hematology 2015;20:397-404. [Crossref] [PubMed]
- Loghavi S, Zuo Z, Ravandi F, et al. Clinical features of de novo acute myeloid leukemia with concurrent DNMT3A, FLT3 and NPM1 mutations. J Hematol Oncol 2014;7:74. [Crossref] [PubMed]
- Piccoli C, Ripoli M, Scrima R, et al. MtDNA mutation associated with mitochondrial dysfunction in megakaryoblastic leukaemic cells. Leukemia 2008;22:1938-41. [Crossref] [PubMed]
- Cui X, Liu F, Wang JQ, et al. Complete sequence analysis of mitochondrial DNA of aplastic anemia patients. Genet Mol Res 2012;11:2130-7. [Crossref] [PubMed]
- Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 2009;361:1058-66. [Crossref] [PubMed]
- Damm F, Bunke T, Thol F, et al. Prognostic implications and molecular associations of NADH dehydrogenase subunit 4 (ND4) mutations in acute myeloid leukemia. Leukemia 2012;26:289-95. [Crossref] [PubMed]
- Shen Y, Zhu YM, Fan X, et al. Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood 2011;118:5593-603. [Crossref] [PubMed]
- Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med 2016;374:2209-21. [Crossref] [PubMed]
- Pastore F, Dufour A, Benthaus T, et al. Combined molecular and clinical prognostic index for relapse and survival in cytogenetically normal acute myeloid leukemia. J Clin Oncol 2014;32:1586-94. [Crossref] [PubMed]
- Wakita S, Yamaguchi H, Ueki T, et al. Complex molecular genetic abnormalities involving three or more genetic mutations are important prognostic factors for acute myeloid leukemia. Leukemia 2016;30:545-54. [Crossref] [PubMed]
- Yan XJ, Xu J, Gu ZH, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet 2011;43:309-15. [Crossref] [PubMed]
- Marková J, Michková P, Burčková K, et al. Prognostic impact of DNMT3A mutations in patients with intermediate cytogenetic risk profile acute myeloid leukemia. Eur J Haematol 2012;88:128-35. [Crossref] [PubMed]
- Nakase K, Bradstock K, Sartor M, et al. Geographic heterogeneity of cellular characteristics of acute myeloid leukemia: a comparative study of Australian and Japanese adult cases. Leukemia 2000;14:163-8. [Crossref] [PubMed]
- Andrieu V, Radford-Weiss I, Troussard X, et al. Molecular detection of t(8;21)/AML1-ETO in AML M1/M2: correlation with cytogenetics, morphology and immunophenotype. Br J Haematol 1996;92:855-65. [Crossref] [PubMed]
- Boissel N, Leroy H, Brethon B, et al. Incidence and prognostic impact of c-Kit, FLT3, and Ras gene mutations in core binding factor acute myeloid leukemia (CBF-AML). Leukemia 2006;20:965-70. [Crossref] [PubMed]


