
Circular RNAs: an emerging type of non-coding RNA and their potential implications in bladder cancer
According to the US National Cancer Institute (NCI; http://www.cancer.gov/ accession October, 2017), bladder cancer (BC) is the 6th most common type of cancer in the Western countries, with ~350,000 new cases/year and a constantly increasing incidence (1). Patients with BC have been commonly stratified according to molecular alterations (i.e., somatic mutations and gene fusions) in FGFR3 and TP53 genes. However, large-scale studies from international consortia (especially from The Cancer Genome Atlas Research Network, Nature 2014) have depicted a more complex picture, reporting the deregulation of a large network of genes (2). Non-coding RNAs, both long and small-RNAs (lncRNAs and miRNAs, respectively)—have been postulated to contribute, at least partially, to the wide deregulation observed in BC. Many striking examples of oncogenic miRNAs and, more recently, lncRNAs—including antisense RNAs [e.g., HOXD-AS1, MDC1-AS1 and ZEB2-AS1 (3-5)]; and the new class of transcribed ultraconserved regions (e.g., T-UCR 8+) (6,7) have been so far reported in the etiology of BC (8).
A recent meta-analysis carried out on PubMed and Google Scholar databases including scientific publications from 1990 to 2016 revealed that at least 35 microRNAs are specifically associated with cellular pathways involved in cellular dedifferentiation, proliferation, and cancer progression, in bladder as well as in other cancer types (9). Most of BC-associated miRs are involved in FGFR3 pathway linked to BC onset and development. The low levels of miR-100 were correlated with low-grade, non-invasive bladder urothelial cancer, and to the upregulation of FGFR3, meaning that—under physiological conditions—miR-100 acts as a tumor suppressor (10). Another example is provided by miR-31 whose reduced expression in urothelial carcinoma of the bladder has been supposed to contribute to BC progression, leading to unfavourable prognosis, overall and progression-free survival (11). In addition, the downregulation of miR-99a, leading to the upregulation of FGFR3, has been frequently linked to low-grade tumors and to a better disease outcome (12). Other miRNAs regulate the TP53 pathway. Among them miR-205 is potentially able to simultaneously target TP53, PTEN (phosphatase and tensin homolog), ERBB3 (a member of the epidermal growth factor receptor family of receptor tyrosine kinases), CDC42 (involved in cell cycle regulation), and the proto-oncogene YES1 (Yamaguchi sarcoma viral oncogene homolog) (13). miR-21 targets many genes involved in cancer onset and progression, including TP53 (14), PTEN and TIMP3 (tissue inhibitor of metalloproteinase 3), both inhibitors of the matrix metalloproteinases, BCL-2 (regulator of apoptosis) (15) and many others; its overexpression is linked to TP53 loss-of-function, commonly observed in high-grade MIBC. The involvement of different microRNAs in the pathogenesis of BC is currently under evaluation and these lists are likely to grow in the next few years due to the wide research efforts in the field.
A relatively new class, i.e., the circular RNAs (circRNAs), has been recently reported to contribute to distinct oncogenic processes in many distinct types of cancers, including BC. CircRNAs lack the canonical 5' cap and 3' poly-A tail and unlike linear RNAs, are characterized by the ends joint together by covalent bonds, which makes them more resistant to exonuclease R and more stable in the cytoplasm. The first observation suggesting that eukaryotic RNAs can exist even in circular form was made more than 30 years ago by electron microscopy (16). CircRNAs’ biogenesis through back splicing differs from the canonical splicing of linear RNAs. Exon circularization depends on the presence of intronic complementary sequences in the flanking regions and alternative formation of inverted Alu repeats that cause alternative circularization. Such a mechanism leads to the generation of multiple circular RNAs from a single gene (17). Nowadays, the application of RNA-Sequencing technology to the study of complex diseases, including cancer, has allowed researchers to discover a large number of circRNAs, transcribed and spliced from exons of protein and non-coding genes. Despite the implementation of sophisticate computational methodologies to discover de novo splice sites, distinguishing bona fide new splicing events from cellular noise is still a challenge (18). Interestingly, this relatively new class of ncRNAs has been found to be abundant in—and evolutionarily conserved across—different species, from Archaea to human (19), and expression studies have reported that they exhibit tissue/developmental-stage-specific characteristics (20,21). However, most of the functions played by these molecules are unknown, despite current literature attributes some peculiar features to circRNAs. One of the most interesting roles of circRNAs relies on the presence of miRNA response elements (MREs) within their sequence. It makes circRNAs able to act as competing endogenous RNAs (ceRNAs) toward other protein-coding transcripts by sponging shared miRNAs. Therefore, interacting with disease-related miRNAs, circRNAs can play important regulatory roles in specific diseases. The most prominent example of this mechanism is the human circRNA CDR1 antisense (CDR1as), that can function as a negative regulator of microRNAs in neuronal tissues due to the presence of 63 conserved binding sites for sequestering miR-7 (22).
Recently, as summarized in Table 1, circRNAs have emerged as critical regulators in tumorigenesis and progression of BC. Zhong and colleagues (24) identified two circRNAs—circFAM169A and circTRIM24—down-regulated and four—circTCF25, circZFR, circPTK2 and circBC048201—up-regulated in bladder carcinoma compared with adjacent non-tumor tissues. Additionally, to further confirm the ability of circRNAs to act as ceRNA they also found that circTCF25 can function as a miRNA sponge by down-regulating miR-103a-3p and miR-107 in cancerous tissue, leading to increased expression of CDK6 gene, ultimately associated with cancer development (24). Li et al. showed that circHIPK3, has a significantly low expression in BC tissues and cell lines and negatively correlates with tumor grade, invasion as well as lymph node metastasis. The authors reported that circHIPK3 contains two critical binding sites for miR-558 and that it can efficiently sponge it inhibiting heparanase expression (HPSE) in BC cells (25).
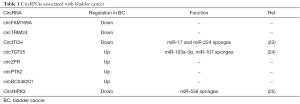
Full table
More recently, a paper published by Yang and colleagues (23) on Molecular Cancer has investigated the role of circ-ITCH in the pathogenesis of BC (23). Circ-ITCH is a well-known circRNA generated from several exons of itchy E3 ubiquitin protein ligase (ITCH). The authors have reported that circ-ITCH is expressed at lower levels in BC compared to the healthy tissue, and that its expression correlates with prognosis of BC patients. In particular, they found that patients with BC expressing low circ-ITCH had a significantly reduced survival rate. Functional assays revealed that cell viability, colony forming ability, migration and invasiveness of BC tumor cell lines forced to over-express circ-ITCH were significantly suppressed. Interestingly, to mechanistically demonstrate its tumor suppressor activity, the authors explored the possibility that circ-ITCH may act as a molecular sponge for oncogenic miRNAs. Then, coupling bioinformatic analysis (to identify MREs) to biotin-coupled probe pull-down assay and FISH they further confirmed the physical interaction of circ-ITCH with miR-17 and miR-224, demonstrating the sponge activity of this circRNA. Interestingly, they also reported that exogenous expression of miR-17 and miR-224 was capable to promote BC cell proliferation and that these miRNAs specifically target p21 and PTEN. Therefore, they postulated that circ-ITCH, sponging miR-17 and miR-224, exerts its anti-tumor activity in BC by modulating the endogenous levels of p21 and PTEN that are natural targets of miR-17 and miR-224. The mechanistic demonstration that circ-ITCH is capable to act as sponge for miR-17 and miR-224, critical post-transcriptional regulators of p21 and PTEN, strongly support circ-ITCH as a new tumor suppressor in BC. This finding may provide clinicians a new potential biomarker, as well as a therapeutic target for the management of BC.
Noteworthy, not all circRNAs can act as “miRNA sponges” (20). Indeed, small sized circRNAs, that appear to be not suitable for miRNA sponging, can be incorporated into exosomes and function as promising biomarkers for cancer diagnosis (26). Intronic circRNAs and exon-intron RNAs, mainly localized in the nucleus, have little enrichment for miRNA binding sites. This type of circRNAs has been reported to regulate parental genes expression levels via specific RNA-RNA interactions (27,28). Other examples, such as circMbl, cricFmn and circDMD, can strongly bind cognate linear transcripts sequestering them from translation and finally leading to a global reduction in protein expression (20,29). This process is also known as “mRNA trap”. Thus, circRNAs differentially expressed in BC cells, beyond acting as “miRNAs sponges”, may differently impact on cancer cell phenotype through distinct mechanisms, most of which still need to be explored.
Conclusions
The further validation of circRNAs as miRNA “sponges” in pathologic conditions makes them excellent candidates for therapeutic purposes, in cancer as well as in other diseases. Thanks to stability, long half-life (up to 48 h), resistance to nucleases, to the capability to act as a ‘reservoir’ preventing miRNA from inhibiting other mRNA targets, to the ability to release miRNAs upon sponging (allowing them to bind their natural target/s), these ncRNAs are a new powerful tool for therapeutic purposes.
Despite the effort to unravel the precise mechanism of action of circRNAs in vitro and in vivo our current knowledge is still rudimentary. However, considering the above described characteristics, as well as their tissue specificity, they have the potential to become new and relevant targets to be added to the collection of available clinical diagnostic markers, and they are also likely to drive new therapeutic strategies for cancer treatment and prevention, including (but not limited to) the BC.
Acknowledgments
Funding: A Cimmino was supported by the Italian Association for Cancer Research (AIRC-MFAG 11510).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Chunlin Ou (Cancer Research Institute of Central South University, Changsha, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.08.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- . Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315-22. [Crossref] [PubMed]
- Li J, Zhuang C, Liu Y, et al. Synthetic tetracycline-controllable shRNA targeting long non-coding RNA HOXD-AS1 inhibits the progression of bladder cancer. J Exp Clin Cancer Res 2016;35:99. [Crossref] [PubMed]
- Xue Y, Ma G, Zhang Z, et al. A novel antisense long noncoding RNA regulates the expression of MDC1 in bladder cancer. Oncotarget 2015;6:484-93. [Crossref] [PubMed]
- Zhuang J, Lu Q, Shen B, et al. TGFbeta1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci Rep 2015;5:11924. [Crossref] [PubMed]
- Terracciano D, Terreri S, de Nigris F, et al. The role of a new class of long noncoding RNAs transcribed from ultraconserved regions in cancer. Biochim Biophys Acta 2017;1868:449-55. [PubMed]
- Olivieri M, Ferro M, Terreri S, et al. Long non-coding RNA containing ultraconserved genomic region 8 promotes bladder cancer tumorigenesis. Oncotarget 2016;7:20636-54. [Crossref] [PubMed]
- Gulia C, Baldassarra S, Signore F, et al. Role of Non-Coding RNAs in the Etiology of Bladder Cancer. Genes (Basel) 2017;8: [Crossref] [PubMed]
- Mitash N, Tiwari S, Agnihotri S, et al. Bladder cancer: Micro RNAs as biomolecules for prognostication and surveillance. Indian J Urol 2017;33:127-33. [Crossref] [PubMed]
- Morais DR, Reis ST, Viana N, et al. The involvement of miR-100 in bladder urothelial carcinogenesis changing the expression levels of mRNA and proteins of genes related to cell proliferation, survival, apoptosis and chromosomal stability. Cancer Cell Int 2014;14:119. [Crossref] [PubMed]
- Wang S, Li Q, Wang K, et al. Decreased expression of microRNA-31 associates with aggressive tumor progression and poor prognosis in patients with bladder cancer. Clin Transl Oncol 2013;15:849-54. [Crossref] [PubMed]
- Catto JW, Miah S, Owen HC, et al. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res 2009;69:8472-81. [Crossref] [PubMed]
- Neely LA, Rieger-Christ KM, Neto BS, et al. A microRNA expression ratio defining the invasive phenotype in bladder tumors. Urol Oncol 2010;28:39-48. [Crossref] [PubMed]
- Catto JW, Alcaraz A, Bjartell AS, et al. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol 2011;59:671-81. [Crossref] [PubMed]
- Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene 2006;25:6170-5. [Crossref] [PubMed]
- Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979;280:339-40. [Crossref] [PubMed]
- Zhang XO, Wang HB, Zhang Y, et al. Complementary sequence-mediated exon circularization. Cell 2014;159:134-47. [Crossref] [PubMed]
- Hooper JE. A survey of software for genome-wide discovery of differential splicing in RNA-Seq data. Hum Genomics 2014;8:3. [Crossref] [PubMed]
- Hou LD, Zhang J. Circular RNAs: An emerging type of RNA in cancer. Int J Immunopathol Pharmacol 2017;30:1-6. [Crossref] [PubMed]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA 2014;20:1829-42. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Yang C, Yuan W, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer 2018;17:19. [Crossref] [PubMed]
- Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep 2016;6:30919. [Crossref] [PubMed]
- Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep 2017;18:1646-59. [Crossref] [PubMed]
- Shohet JM, Ghosh R, Coarfa C, et al. A genome-wide search for promoters that respond to increased MYCN reveals both new oncogenic and tumor suppressor microRNAs associated with aggressive neuroblastoma. Cancer Res 2011;71:3841-51. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. [Crossref] [PubMed]
- Jiang G, Wu AD, Huang C, et al. Isorhapontigenin (ISO) Inhibits Invasive Bladder Cancer Formation In Vivo and Human Bladder Cancer Invasion In Vitro by Targeting STAT1/FOXO1 Axis. Cancer Prev Res (Phila) 2016;9:567-80. [Crossref] [PubMed]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55-66. [Crossref] [PubMed]


