The construction and analysis of gene co-expression network of differentially expressed genes identifies potential biomarkers in thyroid cancer
Introduction
Over the past 4 decades, the incidence of thyroid cancer has been increasing steadily in United States, with 14.42 cases per 100,000 persons per year in 2010–2013 (1). The most common histologic types were papillary thyroid cancer (PTC) (84%) and follicular thyroid cancer (11%). Thyroid cancer mortality rate rose by 1.1% annually from 0.40 per 100,000 person-years in 1994–1997 and to 0.46 per 100,000 person-years in 2010–2013 (1). Therefore, characterization of the molecular pathogenesis of thyroid cancer is critical to identifying key biomarkers and effective therapeutic targets for the disease.
Numerous studies have been conducted to characterize the genomic and transcriptomic landscapes of thyroid cancer. The Cancer Genome Atlas (TCGA) applied MutSig to 496 paired tumor/normal samples and found many frequently mutated driver genes in thyroid cancer, such as BRAF, NRAS, HRAS, EIF1AX and KRAS (2). The study confirmed that PTCs are driven primarily by mutations in one of two cancer-associated genes: BRAF(V600E) or RAS. Yoo et al. classified thyroid tumors into three molecular subtypes, including BRAF-like, RAS-like, and Non-BRAF-Non-RAS based on gene expression profiles. BRAF-like patients showed higher frequency of lymph node metastasis or extrathyroidal extension, while patients in the RAS-like group or Non-BRAF-Non-RAS group showed less or no lymph node metastasis or extrathyroidal extension (3). Costa et al. identified a new gene fusion WNK1-B4GALNT3 that was correlated with B4GALNT3 overexpression in a cohort of 18 PTC patients using RNA-Sequencing (4).
The weighted correlation network analysis (WGCNA) package comprises a series of R functions to perform a variety of weighted correlation network analyses. WGCNA detects co-expression modules of highly correlated genes and associates interested modules with clinical traits, providing insights into signaling networks that may be responsible for phenotypic traits (5). In this study, we performed a genome-wide investigation of differentially expressed genes (DEGs) in 57 pairs of thyroid cancer and normal thyroid tissues. WGCNA was applied to build co-expression modules using the expression data of DEGs in thyroid cancer. Module-trait associations were analyzed using the correlation between the module eigengene and clinical traits. Gene ontology (GO) enrichment analysis was performed on the genes and the hub genes were identified in the modules of interest (6). These findings may be of importance in evaluating the malignant potential and clinical outcomes of thyroid cancer patients.
Methods
DEG and principal component analyses
Among 568 thyroid cancer samples in TCGA, only 57 samples had RNA-sequencing expression data of paired normal thyroid tissues. Therefore, raw count data of 57 thyroid cancer and paired normal thyroid samples were retrieved from the TCGA database at the Broad Institute for DEGs analysis (7) (http://firebrowse.org/?cohort=THCA). In order to further validate DEGs, we obtained RNA-seq expression data of 8 pairs of thyroid cancer and normal thyroid tissues from Gene Expression Omnibus (GEO) (GSE63511) (8). The R package of DEseq2 was used to characterize the expression profiles in 57 thyroid cancer and paired normal tissues (9). If genes showed false discovery rates (FDR) smaller than 0.05 and absolute log2fold-changes greater than 2, they were considered as DEGs. Next, principal component analysis (PCA) was conducted to evaluate whether DEGs could distinguish thyroid cancer from normal tissues.
GO enrichment analysis
To analyze the functional enrichment of DEGs and genes in co-expression modules, GO (6)-enrichment analysis was performed for all the DEGs and genes in co-expression modules with the online tool of GO (http://geneontology.org/). If the cutoff of FDR adjusted P value was smaller than 0.05, the enrichment of GO terms was considered to be significant.
Construction of co-expression network in thyroid cancer
Soft-thresholding power values were screened out in the construction of co-expression modules by the WGCNA algorithm (5). The gradient method was applied to evaluate the scale-free fit index and the mean connectivity degree of different co-expression modules with power values ranging from 1 to 20. The optimal power value was determined when the scale-free fit index was above 0.8, and then the construction of co-expression modules was performed by the WGCNA algorithm in R3.2.0. The minimum number of genes was set as 10 to ensure the high reliability of the results. The corresponding genetic information of co-expression modules was also extracted.
Characterization of module-trait relationships in thyroid cancer
Module-trait associations were analyzed using the correlation between the module eigengenes and clinical phenotypes, which enabled the identification of co-expression modules with significant correlation with the phenotype. For each expression profile, gene significance (GS) was calculated as the absolute value of the correlation between expression profile and each trait; module membership (MM) was defined as the correlation of expression profile and each module eigengene (10). Using the GS and MM measures, genes that have a high significance for each trait as well as high MM in interesting modules could be identified. The intramodular connectivity was computed for each gene by summing the connection strengths with other module genes and dividing this number by the maximum intramodular connectivity. The genes with maximum intramodular connectivity were regarded as intramodular hub genes (11). The interested modules and hub genes were visualized by Integrative Visual Analysis Tool for Biological Networks and Pathways (VisANT) (12) software.
Results
DEGs analysis
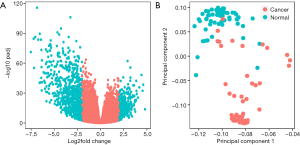
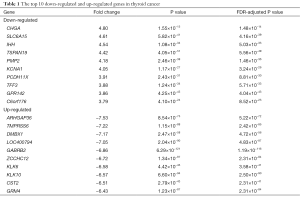
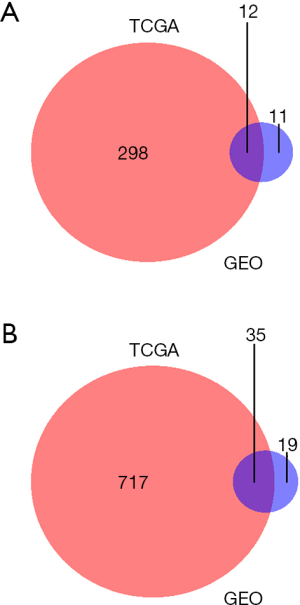
The 57 thyroid cancer samples comprise 48 papillary, 6 follicular and 3 tall cell thyroid cancer samples. Twenty nine cancer samples had BRAFV600E mutations, while 28 samples were wild-type. Six and 51 cancer samples were RAS-mutant or wild-type respectively. Raw count data of thyroid cancer and normal samples were obtained from TCGA. DEGs were determined between 57 pairs of thyroid cancer and normal tissues with DEseq2 package in R. Overall, DEseq2 found 750 up-regulated and 296 down-regulated genes (Figure 1A). ARHGAP36, TMPRSS6, DMBX1, LOC400794, GABRB2, ZCCHC12, KLK6, KLK10, CST2 and GRM4 were the top-ranking up-regulated genes, while CHGA, SLC6A15, IHH, TSPAN19, PMP2, KCNA1, PCDH11X, TFF3, GPR142 and C6orf176 were the top-ranking down-regulated genes in thyroid cancer (Table 1). In order to confirm the DEGs in thyroid cancer, we obtained RNA-seq data of 8 pairs of thyroid cancer and normal thyroid tissues from GEO. 77 genes were determined as DEGs by DESeq2, 64.81% (35/54) up-regulated and 52.17% (12/23) down-regulated genes were overlapped with the DEGs obtained from 57 pairs of thyroid cancer and normal counterparts (Figure S1). Next, PCA was conducted to assess whether DEGs could distinguish thyroid cancer from normal tissues. Cancer and normal tissues were aggregated to the lower and upper sides respectively, demonstrating that DEGs were able to classify the samples into two distinct subsets (Figure 1B).
Full table
GO term enrichment analysis
The enrichment of GO terms was analyzed for 750 up-regulated and 296 down-regulated genes on the home page of GO. GO term enrichment analysis showed down-regulated genes were significantly enriched in 136 biological processes, up-regulated genes in 281 biological processes (FDR adjusted P value <0.05). The main GO biological process terms for down-regulated genes showed a wide variety of functional processes ranging from cellular developmental process, cell differentiation, single-multicellular organismal process, cell development and regulation of system process. While the primary GO terms for up-regulated genes were related to various cellular processes, such as negative regulation of endopeptidase activity, cell-cell adhesion, epidermis development, extracellular matrix organization, regulation of protein phosphorylation, regulation of signal transduction, regulation of cell proliferation and a variety of metabolic processes.
Construction of co-expression modules in thyroid cancer
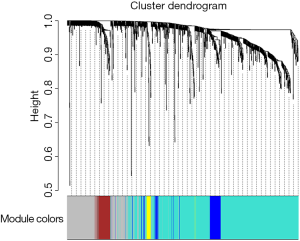
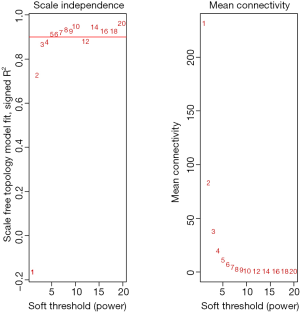
Among the 568 thyroid cancer samples in TCGA, 501 had both RNA-sequencing expression and clinical traits data. Therefore, expression values of 1,046 genes in the 501 samples of thyroid cancer were used to construct the co-expression modules by the WGCNA package. One of the most important parameters was the soft-thresholding power value, which mainly affected the scale-free fit index and the average connectivity degree of co-expression modules. Firstly, the power value was screened out (Figure S2). When the power value was equal to seven, the scale-free fit index was larger than 0.8 and the mean connectivity degree was higher. Therefore, the soft-thresholding power value used to construct co-expression modules and the WGCNA identified five distinct gene co-expression modules which are shown in different colors (Figure 2). These numbers of genes in the five modules differed largely, with 608, 293, 70, 57 and 19 genes for the turquoise, grey, blue, brown and yellow modules respectively (Figure 2).
Module-trait association analysis in thyroid cancer
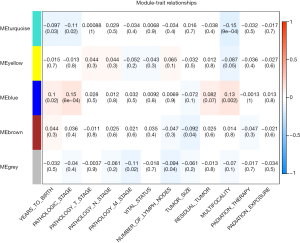
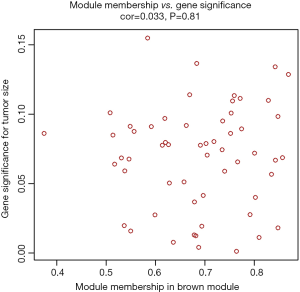
The clinical traits data were obtained from the TCGA database. Module-trait associations were analyzed with the correlation between module eigengene and clinical traits. As showed in the Figure 3, the turquoise module was negatively correlated with patients’ age, cancer stage and multifocality (P value <0.05 for all cases, Figures 2 and 3). In contrast, the blue module was significantly positively associated with patients’ age, cancer stage and multifocality (P value <0.05 for all cases, Figures 2 and 3). The brown module was negatively correlated with tumor size (P value =0.04, Figures 2 and 3). Then, we plotted scatterplots of GS for patients’ age, cancer stage, multifocality and tumor size vs. MM in the blue, turquoise and brown modules. The GS for patients’ age, cancer stage and multifocality was positively correlated with MM in the blue and turquoise modules (P value <0.05 for all cases, Figure 4). However, the GS for tumor size showed non-significant correlation with MM in the brown module (P=0.81, Figure S3).
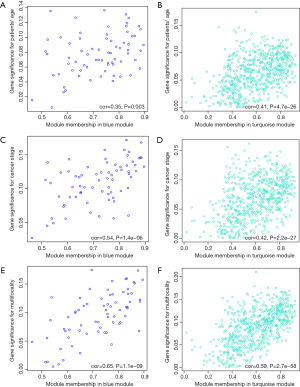
Functional enrichment analysis of genes in the interested modules
GO enrichment analysis was performed on the genes in the turquoise, blue and brown modules. There was large difference in the biological processes in which the three modules were enriched. Genes in turquoise module were significantly over-represented in 171 GO terms (FDR adjusted P value <0.05), such as cell migration (GO:0016477), regulation of cell proliferation (GO:0042127), regulation of cell differentiation (GO:0045595), regulation of cellular protein metabolic process (GO:0032268), signal transduction (GO:0007165). While genes in the turquoise module were significantly under-represented in 42 GO terms, such as gene expression (GO:0010467), DNA repair (GO:0006281), DNA metabolic process (GO:0006259) and cellular response to DNA damage stimulus (GO:0006974). However, no significant enrichment of GO terms was found for the genes in the blue and brown modules.
Visualization of the interested modules and hub genes
The modules were constructed using the VisANT software. The genes with largest intramodular connectivity were considered as intramodular hub genes. The hub genes in the turquoise and blue modules were bold with red in Figure 5. SERPINA1 and MRO were the hub genes in the turquoise and blue modules respectively (Figure 5).

Discussion
RNA-seq analyses have previously been used to predict therapeutic biomarkers and to identify gene expression patterns associated with clinical traits in thyroid cancer (13,14). This study presented a large number of DEGs between 57 pairs of thyroid cancer and normal tissues. By comparing the DEGs to the tumor associated gene and tumor suppressor databases (15), five known oncogenes were up-regulated, including ELF3, HMGA2, LCN2, MET and RUNX2; by contrast, 10 TSGs were found down-regulated, including EGR2, FOXA2, GPC3, IGFBPL1, LRP1B, MT1G, NR4A3, PROX1, TMEFF2 and WNT11. Additionally, some new DEGs were discovered, such as ARHGAP36, TMPRSS6, DMBX1, CHGA and SLC6A15, which have not been reported previously and their functions remain unknown in thyroid cancer.
WGCNA is a method frequently used to explore the co-expression modules of highly correlated genes. Genes in the same module were considered to be related with each other in function. Therefore, the analysis allowing for identification of biologically-relevant modules and hub genes that may eventually serve as biomarkers for detection or treatment (5). In this study, a total of five co-expression modules were constructed by the 1,046 genes from the 57 pairs of thyroid cancer and normal samples by the WGCNA method. We identified the blue and turquoise co-expression modules that relate to clinical traits (patient’s age, cancer stage and multifocality, Figures 2-4). The result of functional enrichment analysis showed that the turquoise module was found to be mainly enriched in cellular processes associated with cell migration, regulation of cell proliferation, regulation of cell differentiation, and regulation of cellular protein metabolic process. While genes in the blue module were not significantly enriched in any GO terms. Therefore, we speculate that turquoise module is the most important module in the cancer stage and multifocality of thyroid cancer.
The turquoise and blue co-expression modules were constructed by the VisANT software and the hub genes SERPINA1 and MRO were identified (Figure 5). The SERPINA1, also known as α1-AntiTrypsin, is a protease inhibitor that can act on a variety of targets such as serine proteases. It has been demonstrated that SERPINA1 expression can be stimulated by E2 in MCF-7 cells, and a high expression of this protein inhibits colony formation (16). SERPINA1 is up-regulated in colorectal cancer (17), glioma (18), cutaneous squamous cell carcinoma (19) and thyroid cancer (20). SERPINA1 has been proposed as a biomarker for various cancers, such as papillary thyroid carcinoma (20), lung cancer (21) and breast carcinoma (22-24). The high expression of SERPINA1 indicates a poor prognosis of high-grade glioma (18) but a favorable clinical outcome in breast cancer patients (25). The hub gene of the blue module is MRO, which is down-regulated in lung cancer cell lines (26). The hub genes in the turquoise and blue modules are either oncogenes or anti-oncogenes. Therefore, we speculate that the hub genes are critical in the cancer stage and multifocality of thyroid cancer.
Conclusions
Taken together, the turquoise module was regarded as the most critical module in the cancer stage and multifocality of thyroid cancer. More importantly, the hub gene SERPINA1 may function as a potential biomarker for thyroid cancer, which also needs much further research.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.09.12). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lim H, Devesa SS, Sosa JA, et al. Trends in thyroid cancer incidence and mortality in the United States, 1974-2013. JAMA 2017;317:1338-48. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Papillary Thyroid Carcinoma. Cell 2014;159:676-90. [Crossref] [PubMed]
- Yoo SK, Lee S, Kim SJ, et al. Comprehensive Analysis of the Transcriptional and Mutational Landscape of Follicular and Papillary Thyroid Cancers. PLoS Genet 2016;12:e1006239 [Crossref] [PubMed]
- Costa V, Esposito R, Ziviello C, et al. New somatic mutations and gene fusion in papillary thyroid carcinoma. Oncotarget 2015;6:11242-51. [Crossref] [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113-20. [Crossref] [PubMed]
- Riesco-Eizaguirre G, Wert-Lamas L, Perales-Paton J, et al. The miR-146b-3p/PAX8/NIS regulatory circuit modulates the differentiation phenotype and function of thyroid cells during carcinogenesis. Cancer Res 2015;75:4119-30. [Crossref] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Shi K, Bing ZT, Cao GQ, et al. Identify the signature genes for diagnose of uveal melanoma by weight gene co-expression network analysis. Int J Ophthalmol 2015;8:269-74. [PubMed]
- Wisniewski N, Cadeiras M, Bondar G, et al. Weighted Gene Coexpression Network Analysis (WGCNA) Modeling of Multiorgan Dysfunction Syndrome after Mechanical Circulatory Support Therapy. J Hear Lung Transplant 2018;32:S223. [Crossref]
- Hu Z, Mellor J, Wu J, et al. VisANT: An online visualization and analysis tool for biological interaction data. BMC Bioinformatics 2004;5:17. [Crossref] [PubMed]
- Qiu J, Zhang W, Xia Q, et al. RNA sequencing identifies crucial genes in papillary thyroid carcinoma (PTC) progression. Exp Mol Pathol 2016;100:151-9. [Crossref] [PubMed]
- Baldan F, Mio C, Allegri L, et al. Identification of tumorigenesis-related mRNAs associated with RNA-binding protein HuR in thyroid cancer cells. Oncotarget 2016;7:63388-407. [Crossref] [PubMed]
- Zhao M, Sun J, Zhao Z. TSGene: A web resource for tumor suppressor genes. Nucleic Acids Res 2013;41:D970-6. [Crossref] [PubMed]
- Finlay TH, Tamir S, Kadner SS, et al. alpha 1-Antitrypsin- and anchorage-independent growth of MCF-7 breast cancer cells. Endocrinology 1993;133:996-1002. [Crossref] [PubMed]
- Pérez-Holanda S, Blanco I, Menéndez M, et al. Serum concentration of alpha-1 antitrypsin is significantly higher in colorectal cancer patients than in healthy controls. BMC Cancer 2014;14:355. [Crossref] [PubMed]
- Ookawa S, Wanibuchi M, Kataoka-Sasaki Y, et al. Digital Polymerase Chain Reaction Quantification of SERPINA1 Predicts Prognosis in High-Grade Glioma. World Neurosurg 2018;111:e783-9. [Crossref] [PubMed]
- Farshchian M, Kivisaari A, Ala-Aho R, et al. Serpin peptidase inhibitor clade a member 1 (SerpinA1) is a novel biomarker for progression of cutaneous squamous cell carcinoma. Am J Pathol 2011;179:1110-9. [Crossref] [PubMed]
- Vierlinger K, Mansfeld MH, Koperek O, et al. Identification of SERPINA1 as single marker for papillary thyroid carcinoma through microarray meta analysis and quantification of its discriminatory power in independent validation. BMC Med Genomics 2011;4:30. [Crossref] [PubMed]
- Topic A, Ljujic M, Nikolic A, et al. Alpha-1-antitrypsin phenotypes and neutrophil elastase gene promoter polymorphisms in lung cancer. Pathol Oncol Res 2011;17:75-80. [Crossref] [PubMed]
- López-Árias E, Aguilar-Lemarroy A, Felipe Jave-Suárez L, et al. Alpha 1-antitrypsin: A novel tumor-associated antigen identified in patients with early-stage breast cancer. Electrophoresis 2012;33:2130-7. [Crossref] [PubMed]
- Doustjalali SR, Yusof R, Yip CH, et al. Aberrant expression of acute-phase reactant proteins in sera and breast lesions of patients with malignant and benign breast tumors. Electrophoresis 2004;25:2392-401. [Crossref] [PubMed]
- Abbott KL, Aoki K, Lim JM, et al. Targeted glycoproteomic identification of biomarkers for human breast carcinoma. J Proteome Res 2008;7:1470-80. [Crossref] [PubMed]
- Chan HJ, Li H, Liu Z, et al. SERPINA1 is a direct estrogen receptor target gene and a predictor of survival in breast cancer patients. Oncotarget 2015;6:25815-27. [Crossref] [PubMed]
- Yanaihara N, Kohno T, Takakura S, et al. Physical and Transcriptional Map of a 311-kb Segment of Chromosome 18q21, a Candidate Lung Tumor Suppressor Locus. Genomics 2001;72:169-79. [Crossref] [PubMed]