
Prognostic and clinicopathological roles of long non-coding RNA XIST in human cancers: a meta-analysis
Introduction
Cancer is currently one of the deadliest diseases known to man, and a major threat to human health. A reported 1,688,780 new cases and 600,920 associated mortalities are estimated to have occurred in the United States in 2017 between January and September (1). Due to diagnosis delays, disease recurrence and chemoresistance, the prognosis of cancer is typically poor and the survival rate is low (2,3). Therefore, current research is focused on identifying potential novel prognostic biomarkers for cancers (4-6).
Long non-coding RNAs (lncRNAs), longer than 200 nucleotides in length, are a class of newly identified non-coding transcripts (7,8). During the past decade, lncRNAs have attracted attention and prompted a series of recent studies due to their regulation of multiple cancer-related cellular processes, including proliferation, invasion, metastasis, angiogenesis, apoptosis and chemoresistance (9-14). This suggests that lncRNAs may turn out to be a promising biomarker for prognosis prediction in cancers (15,16).
X-inactive specific transcript (XIST), derived from the XIST gene, is a newly identified lncRNA which plays a key regulatory role in X inactivation during female mammalian development (17). Increasing studies have revealed that the expression of lncRNA XIST not only plays an important role in the differentiation, proliferation, and genome maintenance of human cells, but it is also involved in cancer cell proliferation, invasion, epithelial-mesenchymal transition (EMT) and metastasis (18). Recently, a growing body of research has shown that XIST is overexpressed in cancer, including non-small-cell lung cancer (19), glioma (20), ovarian cancer (21), and hepatocellular carcinoma (22). Despite this evidence, few meta-analyses have been adopted to determine the association between XIST and cancer prognosis. Therefore, we designed this meta-analysis to investigate the prognostic value of XIST in various cancers.
Methods
Data collection
This meta-analysis was conducted in-line with the PRISMA guidelines (23). Literature from the PubMed, Cochrane Library, EMBASE, Web of Science, CNKI and Wan Fang databases were screened for articles published up to August 2017. The search included the following key words: “XIST”, “long non-coding RNA XIST”, “cancer”, “tumor”, “carcinoma” and “neoplasm”.
Inclusion and exclusion criteria
The inclusion criteria were as follows: (I) a study of any type of human cancer; (II) the association of XIST expression levels with overall survival (OS), recurrence-free survival (RFS), or description of clinicopathological characteristics; (III) the method of detecting lncRNA XIST restricted to reverse transcription quantitative polymerase chain reaction; (IV) patients categorized into low and high groups; (V) sufficient data available for calculating the hazard ratios (HR) and corresponding 95% confidence intervals (CI). The exclusion criteria were as follows: (I) case reports, reviews, meta-analyses, and duplicate publications; (II) studies not presenting data with relevant values; (III) studies only focusing on the molecular mechanisms of lncRNA XIST.
Data extraction
Two reviewers extracted the data from each enrolled publication, including the first author’s name, publication year, country of origin, type of cancer, number of cancer stage, detection method and outcomes. When the HR and 95% CI for survival analysis could not be obtained, we calculated the HRs and the 95% CIs using Kaplan-Meier curves and observed data based on the methods outlined by Tierney et al. (24). The Newcastle-Ottawa scale (NOS) for cohort studies was also conducted to assess the quality of involved studies by 2 authors.
Statistical analysis
HRs and 95% CIs were used to evaluate the association between XIST and cancer prognosis. Furthermore, odds ratios (ORs) and 95% CIs were calculated to determine the associations between XIST and the clinicopathological characteristics detailed in the included articles. The Cochrane Q-test and P values were utilized to assess the heterogeneity. If significant heterogeneity was considered present (I2 ≥50% and P≤0.1), the random-effects model was selected to calculate the pooled HRs or ORs; otherwise, the fixed-effects model was applied. The Begg’s test was used to test publication bias. Statistical analysis was carried out using STATA version 12.0 (Stata Corporation, College Station, TX, USA). P<0.05 was considered statistical significance.
Results
Characteristics of included studies
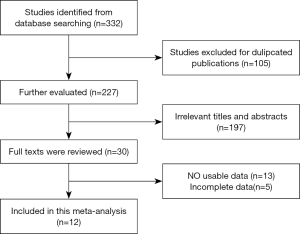
We retrieved a total of 332 articles based on the aforementioned search strategy and removed 105 duplicates, as shown in Figure 1. Following title and abstract screening, 30 accessible full-text articles were identified. A further 18 of those 30 articles were excluded due to lack of data or incomplete data. Finally, 12 studies, comprising a total of 841 patients, were chosen for the final analyses (25-36). Table 1 summarizes the statistical analyses of the 12 studies.
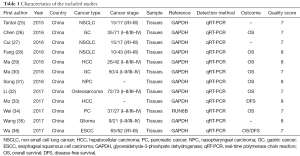
Full table
All the studies included are from China. There were 8 different types of cancer detected, including 2 cases of gastric cancer, 1 case of nasopharyngeal carcinoma, one case of osteosarcoma, 1 case of pancreatic cancer, 2 cases of hepatocellular carcinoma, 1 case of glioma, 3 cases of non-small cell lung cancer and 1 case of esophageal squamous cell carcinoma.
Association between XIST and prognosis
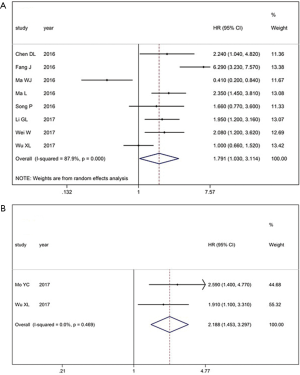
There was a total of 8 studies including the reported data from 841 patients on XIST expression and OS. Due to heterogeneity (I2 =87.9%, P<0.001), the random-effects model was selected to evaluate the pooled HRs with 95% CIs. Compared with the low XIST expression group, the high XIST expression group had a statistically significantly shorter OS (HR =1.791, 95% CI: 1.030–3.114, P<0.001; Figure 2A). Next, the prognostic role of XIST in RFS was evaluated in 2 studies. We observed that the high XIST group had a statistically significantly shorter RFS (HR =2.188, 95% CI: 1.453–3.297, P<0.469, Figure 2B).
Association between XIST and clinicopathological parameters
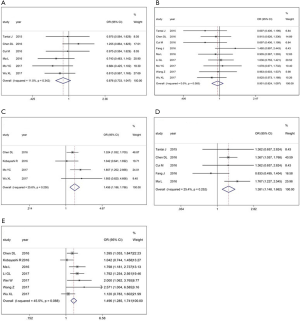
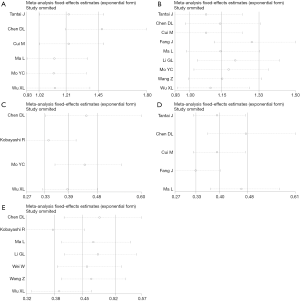
First, the results from 4 studies demonstrated that there was a positive correlation between XIST expression and tumor size (OR =1.456, 95% CI: 1.186–1.786; P=0.258; Figure 3). Additionally, the results from 5 studies showed that the increase in XIST expression was significantly related with lymph node metastasis (OR =1.381, 95% CI: 1.148–1.662; P=0.252; Figure 3D). Findings also showed that an increase in XIST expression was significantly associated with advanced TNM stage (OR =1.496, 95% CI: 1.285–1.741; P<0.088; Figure 3E). However, no significant correlation was found with age and gender (age: Figure 3A; gender: Figure 3B).
Publication bias and sensitivity analysis
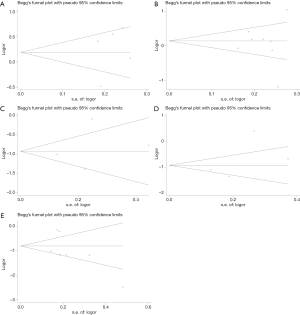
The Begg’s test was applied to assess publication bias in these studies, and no obvious publication bias in the studies was found (Figure 4). A sensitivity analysis was executed to determine whether any individual study affected the overall results. We found these results were not stable and more studies need to be involved in the research to validate the results in the future (Figure 5).
Discussion
Recently, evidence from a growing number of publications have suggested that lncRNAs were dysregulated in various types of cancer and served key roles in the progression of various cancers (37-40). Moreover, lncRNAs may also be considered as promising biomarkers for estimating the prognosis of different cancers (41-43).
XIST, a lncRNA derived from the XIST gene, is a key regulatory factor of X inactivation in mammals (44,45). A series of studies have reported that XIST expression levels in cancer tissues are markedly high compared to those in the adjacent normal tissues (28,34). Accumulating evidence indicates that the expression levels of XIST affect cell proliferation, apoptosis, invasion and metastasis in various cancers (30,32,46). In one study, XIST was identified as an oncogenic lncRNA by competitively binding to miRNAs, thereby modulating the de-repression of miRNA targets (47). In addition to this, Wu et al. (36) observed that XIST contributed to the development of malignancies such as esophageal squamous cell carcinoma through competitive binding with miR-101 to regulate EZH2 expression. Moreover, Fang et al. (28) discovered that XIST knockdown could inhibit cell proliferation, migration and invasion via epigenetically silencing KLF2 expression in non-small cell lung cancer. From a prognostic perspective, high XIST expression was reported to be closely identified with poor prognosis in several types of cancers, including gastric cancer, pancreatic cancer, glioma, osteosarcoma, nasopharyngeal carcinoma, non-small cell lung cancer and hepatocellular carcinoma.
The purpose of this paper was to determine the prognostic roles of lncRNA XIST in human cancers. Based on the results, high XIST expression was proven to be significantly correlated with poor OS or RFS in patients with cancer. These results showed that XIST could act as a prognostic biomarker for OS or RFS in cancer patients. In addition, our findings demonstrated that high XIST expression was positively associated with tumor size, lymph node metastasis, and advanced TNM stage. Moreover, statistical analysis revealed that no obvious associations were found between high XIST expression and age or gender.
However, there were several limitations to the meta-analysis. Firstly, all the studies were from China, thus the results may not be applicable globally. Secondly, the cut-off level for dividing patients into high and low XIST expression groups varied among the different studies. Finally, available data were not deemed sufficient to validate the relationship between XIST and clinicopathological parameters, which requires further research.
Conclusions
In summary, our data illustrates that XIST is a novel prognostic biomarker for patients with cancer. However, further large-scale studies on specific cancer types are required to validate the prognostic roles of XIST in cancer.
Acknowledgments
Funding: This work was supported in part by the Natural Science Foundation of Shandong Province (No. ZR2015AL014) and the Fundamental Research Funds for the Central Universities (No. 15CX02064A).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.12.13). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Ma PJ, Guan QK, Meng L, et al. Long non-coding RNA TUG1 as a potential prognostic biomarker in human cancers: a meta-analysis. Oncotarget 2017;8:62454-62. [PubMed]
- Elkholi R, Renault TT, Serasinghe MN, et al. Putting the pieces together: How is the mitochondrial pathway of apoptosis regulated in cancer and chemotherapy? Cancer Metab 2014;2:16. [Crossref] [PubMed]
- Kojima T, Kawai K, Miyazaki J, et al. Biomarkers for precision medicine in bladder cancer. Int J Clin Oncol 2017;22:207-13. [Crossref] [PubMed]
- Ribeiro IP, Barroso L, Marques F, et al. Early detection and personalized treatment in oral cancer: the impact of omics approaches. Mol Cytogenet 2016;9:85. [Crossref] [PubMed]
- Ray K. Pancreatic cancer: Biomarkers for the early detection of PDAC. Nat Rev Gastroenterol Hepatol 2017;14:504-5. [Crossref] [PubMed]
- Zhuo C, Jiang R, Lin X, et al. LncRNA H19 inhibits autophagy by epigenetically silencing of DIRAS3 in diabetic cardiomyopathy. Oncotarget 2017;8:1429-37. [Crossref] [PubMed]
- Johnsson P, Lipovich L, Grander D, et al. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta 2014;1840:1063-71. [Crossref] [PubMed]
- Zhang J, Gao Y. CCAT-1 promotes proliferation and inhibits apoptosis of cervical cancer cells via the Wnt signaling pathway. Oncotarget 2017;8:68059-70. [PubMed]
- Chen W, Zhu H, Yin L, et al. lncRNA-PVT1 Facilitates Invasion Through Upregulation of MMP9 in Nonsmall Cell Lung Cancer Cell. DNA Cell Biol 2017;36:787-93. [Crossref] [PubMed]
- Li Z, Dong M, Fan D, et al. LncRNA ANCR down-regulation promotes TGF-beta-induced EMT and metastasis in breast cancer. Oncotarget 2017;8:67329-43. [PubMed]
- Wang Y, Liang T, Wang Y, et al. Long non-coding RNA AK093407 promotes proliferation and inhibits apoptosis of human osteosarcoma cells via STAT3 activation. Am J Cancer Res 2017;7:892-902. [PubMed]
- Pan Y, Chen J, Tao L, et al. Long noncoding RNA ROR regulates chemoresistance in docetaxel-resistant lung adenocarcinoma cells via epithelial mesenchymal transition pathway. Oncotarget 2017;8:33144-58. [PubMed]
- Yu H, Xue Y, Wang P, et al. Knockdown of long non-coding RNA XIST increases blood-tumor barrier permeability and inhibits glioma angiogenesis by targeting miR-137. Oncogenesis 2017;6:e303 [Crossref] [PubMed]
- Liao M, Li B, Zhang S, et al. Relationship between LINC00341 expression and cancer prognosis. Oncotarget 2017;8:15283-93. [Crossref] [PubMed]
- Martini P, Paracchini L, Caratti G, et al. lncRNAs as Novel Indicators of Patients' Prognosis in Stage I Epithelial Ovarian Cancer: A Retrospective and Multicentric Study. Clin Cancer Res 2017;23:2356-66. [Crossref] [PubMed]
- Engreitz JM, Pandya-Jones A, McDonel P, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 2013;341:1237973 [Crossref] [PubMed]
- Yang Z, Jiang X, Jiang X, et al. X-inactive-specific transcript: A long noncoding RNA with complex roles in human cancers. Gene 2018;679:28-35. [Crossref] [PubMed]
- Wang H, Shen Q, Zhang X, et al. The Long Non-Coding RNA XIST Controls Non-Small Cell Lung Cancer Proliferation and Invasion by Modulating miR-186-5p. Cell Physiol Biochem 2017;41:2221-9. [Crossref] [PubMed]
- Cheng Z, Li Z, Ma K, et al. Long Non-coding RNA XIST Promotes Glioma Tumorigenicity and Angiogenesis by Acting as a Molecular Sponge of miR-429. J Cancer 2017;8:4106-16. [Crossref] [PubMed]
- Ren C, Li X, Wang T, et al. Functions and Mechanisms of Long Noncoding RNAs in Ovarian Cancer. Int J Gynecol Cancer 2015;25:566-9. [Crossref] [PubMed]
- Zhuang LK, Yang YT, Ma X, et al. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis 2016;7:e2203 [Crossref] [PubMed]
- Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J Clin Epidemiol 2009;62:e1-34. [Crossref] [PubMed]
- Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. [Crossref] [PubMed]
- Tantai J, Hu D, Yang Y, et al. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Pathol 2015;8:7887-95. [PubMed]
- Chen DL, Ju HQ, Lu YX, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res 2016;35:142. [Crossref] [PubMed]
- Cui M, Huo X, Chen B. The analysis on the significance of combined detection of serum long non-coding RNA XIST and HIF1A-AS1 in non-small cell lung cancer screening. Clin Res Pract 2016;1:1-3.
- Fang J, Sun CC, Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun 2016;478:811-7. [Crossref] [PubMed]
- Ma W, Wang H, Jing W, et al. Downregulation of long non-coding RNAs JPX and XIST is associated with the prognosis of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol 2017;41:163-70. [Crossref] [PubMed]
- Ma L, Zhou Y, Luo X, et al. Long non-coding RNA XIST promotes cell growth and invasion through regulating miR-497/MACC1 axis in gastric cancer. Oncotarget 2017;8:4125-35. [PubMed]
- Song P, Ye LF, Zhang C, et al. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene 2016;592:8-14. [Crossref] [PubMed]
- Li GL, Wu YX, Li YM, et al. High expression of long non-coding RNA XIST in osteosarcoma is associated with cell proliferation and poor prognosis. Eur Rev Med Pharmacol Sci 2017;21:2829-34. [PubMed]
- Mo Y, Lu Y, Wang P, et al. Long non-coding RNA XIST promotes cell growth by regulating miR-139-5p/PDK1/AKT axis in hepatocellular carcinoma. Tumour Biol 2017;39:1010428317690999 [Crossref] [PubMed]
- Wei W, Liu Y, Lu Y, et al. LncRNA XIST Promotes Pancreatic Cancer Proliferation Through miR-133a/EGFR. J Cell Biochem 2017;118:3349-58. [Crossref] [PubMed]
- Wang Z, Yuan J, Li L, et al. Long non-coding RNA XIST exerts oncogenic functions in human glioma by targeting miR-137. Am J Transl Res 2017;9:1845-55. [PubMed]
- Wu X, Dinglin X, Wang X, et al. Long noncoding RNA XIST promotes malignancies of esophageal squamous cell carcinoma via regulation of miR-101/EZH2. Oncotarget 2017;8:76015-28. [PubMed]
- Du M, Wang W, Jin H, et al. The association analysis of lncRNA HOTAIR genetic variants and gastric cancer risk in a Chinese population. Oncotarget 2015;6:31255-62. [Crossref] [PubMed]
- Wang D, Ding L, Wang L, et al. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget 2015;6:41045-55. [Crossref] [PubMed]
- Chakravarty D, Sboner A, Nair SS, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun 2014;5:5383. [Crossref] [PubMed]
- Loewen G, Jayawickramarajah J, Zhuo Y, et al. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol 2014;7:90. [Crossref] [PubMed]
- Huang C, Liu S, Wang H, et al. LncRNA PVT1 overexpression is a poor prognostic biomarker and regulates migration and invasion in small cell lung cancer. Am J Transl Res 2016;8:5025-34. [PubMed]
- Yang F, Lv SX, Lv L, et al. Identification of lncRNA FAM83H-AS1 as a novel prognostic marker in luminal subtype breast cancer. Onco Targets Ther 2016;9:7039-45. [Crossref] [PubMed]
- Milevskiy MJ, Al-Ejeh F, Saunus JM, et al. Long-range regulators of the lncRNA HOTAIR enhance its prognostic potential in breast cancer. Hum Mol Genet 2016;25:3269-83. [Crossref] [PubMed]
- Brown CJ, Hendrich BD, Rupert JL, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 1992;71:527-42. [Crossref] [PubMed]
- Brown CJ, Ballabio A, Rupert JL, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 1991;349:38-44. [Crossref] [PubMed]
- Tang Y, He R, An J, et al. lncRNA XIST interacts with miR-140 to modulate lung cancer growth by targeting iASPP. Oncol Rep 2017;38:941-8. [Crossref] [PubMed]
- Chang S, Chen B, Wang X, et al. Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer 2017;17:248. [Crossref] [PubMed]


