Tamoxifen side effects: pharmacogenetic and clinical approach in Mexican mestizos
Introduction
Tamoxifen has been widely used as an adjuvant treatment in patients diagnosed with estrogen receptor-positive breast cancer (ER+ breast cancer) (1). A selective estrogen receptor modulator (SERM), it is a prodrug extensively metabolized by enzymes belonging to the cytochrome P450 superfamily, particularly CYP2D6. Metabolism generates two main active metabolites: 4-hydroxytamoxifen and endoxifen (2). The CYP2D6 gene is highly polymorphic, attaining at least 130 single-nucleotide polymorphisms (SNP) that define >100 allelic variants (3). These are translated into four genetic phenotypes (GP): genetic poor metabolizer (gPM); genetic intermediate metabolizer (gIM); genetic normal metabolizer (gNM); and genetic ultra-rapid metabolizer (gUM) (4,5).
Although CYP2D6 is clearly involved in tamoxifen biotransformation, its association with therapeutic efficacy is still debated, with various opinions for (6-8) and against (9,10).
Approved for clinical applications in 1970, tamoxifen has been shown to reduce recurrence indices and improve five-year disease-free survival rates (11). Results from the Adjuvant Tamoxifen Longer Against Shorter trial highlight the need for a possible extension to a ten-year treatment period (12). Several tamoxifen side effects (TSE) are generated by its pro- and anti-estrogenic activity (13). These TSE include hot flashes (HF), headache, arthralgia, cramps, retinopathy, vomiting, dizziness, night sweats, myelosuppression, thromboembolism, and endometrial cancer (14). The most frequent symptom in a diverse patient population has been HF, and it has consequently been proposed as an indicator of tamoxifen therapeutic efficacy, although conclusions have varied (8,9,15).
Some clinical studies have evaluated the predictive potential of CYP2D6 for TSE occurrence, but results have been contradictory. For instance, gPM patients are reported to have a lower HF incidence (8), which is plausible since CYP2D6 is responsible for tamoxifen activation to endoxifen. In another study, gUM patients developed a higher number of TSE than the other phenotypes (16). Other authors, however, have not identified any association between CYP2D6 and TSE (9,15). In addition, variable results have been observed in evaluations of potential associations between HF and CYP2D6 (8,9,17). Much of this interstudy inconsistency can be attributed to lack of methodological rigor; for example, some studies have not considered CYP2D6 inhibitor use and therapeutic compliance (18).
Decision-making for patients under adjuvant treatment would be better informed with more pharmacogenetic data on widely used therapies. However, substantial pharmacogenetic data for drug metabolizing enzymes are only available for certain populations, and is lacking or poor for many others. In Mexico, some studies have been done to redress this shortcoming though none has properly tested their clinical impact. Instead, they were designed to describe the most prevalent alleles in some CYP genes, including nonfunctional variants related with the gPM status such as: CYP2C19*2 (9.20%) and *3 (0.10%) (19); CYP2C9*2, *3, and *6 with frequencies of 7.00%, 1.50%, and 0.50%, respectively (20); and CYP2D6*3 (0.90–1.50%), CYP2D6*4 (11.00–13.00%), and *5 (1.00–2.00%) (21,22).
This diversity in nonfunctional variants in various drug metabolizing enzymes could be attributed to the ethnic heterogeneity of Mexico’s population which is the result of intermixing of various Amerindian populations with predominantly European emigrants, creating what are known as Mestizo populations (23). Genetic variation in Mexico responds to these Amerindian and European ancestries, and exhibits some degree of grouping based on geography; differences have been reported between regions in the north (e.g., Sonora, Nuevo Leon, and Durango) and south (e.g., Chiapas, Quintana Roo, and Yucatan) (23). The state of Yucatan is notable in this respect because its Mestizo group has been heavily influenced by the Maya, a major Amerindian population in the region (23). This substantial contribution from a single group may explain the genetic diversity observed in Mestizos from Yucatan, and may have produced inter-individual variability in genes such as CYP2D6 (23,24).
Genetic heterogeneity may be associated with variability in and presentation of TSE. To our knowledge, no research that complies with international recommendations for non-prospective studies, in such a poorly represented group in literature as the Mestizos from Yucatan, Mexico, has been reported on the predictive value of CYP2D6 (or other clinical variables) for TSE (18). Although the influence of absent/decreased activity in CYP2D6 genotypes has been extensively explored to test the efficacy of tamoxifen treatment (mostly in Caucasian and Asian groups), very limited attention has been given the relationship between CYP2D6 polymorphisms and TSE (25). The present study aim was to analyze CYP2D6 genotypes and phenotypes, and clinical characteristics, as potential predictors of TSE in highly adherent ER+ breast cancer Mestizo patients from Yucatan, Mexico, without CYP2D6 inhibitor intake.
Methods
Study design and patients
A cross-sectional, analytical study was done of ER+ breast cancer women from Merida, Yucatan, Mexico, under adjuvant tamoxifen (20 mg/day) treatment. Considering a 10% gPM prevalence in Mexican Mestizos (22,26), and assuming a type 1 error (α=0.05) and 5% accuracy; the minimum estimated sample size was 71 patients (27). Study design power to detect significant pharmacogenetic associations was estimated in 60% using the QUANTO software (28) with the following conditions: sample size =71, type I error of 0.007 (after Bonferroni correction, taking into account the four studied polymorphic loci); and a dominant genetic model.
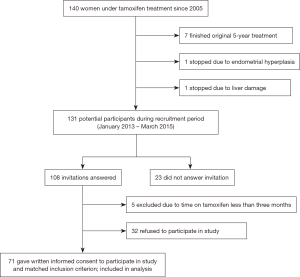
Inclusion criteria included high patient adherence to tamoxifen treatment. Exclusion criteria included CYP2D6 inhibitor/inductor therapy (Table S1), and/or a tamoxifen treatment period of less than three months (18). Participant recruitment resulted in a total of 71 women consenting to participate (Figure 1). None was metastatic at the time of recruitment (January 2013 to March 2015). All participants were seen at the Oncology Department of the Specialty Medical Unit (Unidad Médica de Alta Especialidad - UMAE), and family practice clinics, of the Mexican Social Security Institute (Instituto Mexicano del Seguro Social - IMSS) in Merida, Yucatan, Mexico.
All procedures complied with the ethical standards of the IMSS National Clinical Research Ethics Committee (R-2013-785-057), the Dr. Hideyo Noguchi Regional Research Center Review Board for Ethical Research with Human Subjects, and the 1964 Helsinki Declaration and its amendments. After explanation of the study objectives, its contribution to improved understanding of tamoxifen metabolism, data confidentiality, and the possible complications of venipuncture, written informed consent was obtained individually from all study participants.
Data collection: demographic, socioeconomic, side effects, and adherence
A questionnaire was applied to participants to collect personal data; height (m); weight (kg); body mass index (BMI, kg/m2); medical history; previous breast cancer treatment; tamoxifen use duration; TSE; and treatment adherence. Adherence was defined as a ≥80% dosage compliance during the previous month as quantified by pill count (29-32). Questionnaire items included yes/no questions on the occurrence of six TSE: HF; headache; cramps; arthralgia; dizziness; and vomiting. Items also addressed TSE severity, which was rated on a Likert-type scale based on the Common Terminology Criteria for Adverse Events of the U.S. National Cancer Institute: 1 (mild), 2 (moderate), and 3 (severe) (33). Patients were instructed to report the TSE only if they had begun to appear after tamoxifen treatment initiation.
Socioeconomic status was assessed with the Graffar’s classification system (34), which evaluates education level, occupation, family income source, and household characteristics.
Menopausal status was assigned as follows: (I) pre-menopausal, having had at least one menstrual period during the prior three months with no changes in regularity during previous year; and (II) post-menopausal, amenorrhea during at least six continuous months (15).
Laboratory procedures—genomic DNA extraction and genotyping
A single 4 mL sample of peripheral blood was collected from each participant for DNA isolation. Samples were kept at 4–8 °C until processed. Genomic DNA was extracted from EDTA-anticoagulated total blood with the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA) following manufacturer instructions. Genotyping of CYP2D6 was done with the TaqMan® Allelic Discrimination Assay (Applied Biosystems, Foster City, CA, USA) using a StepOne device (Applied Biosystems) and following supplier specifications. Based on the most frequent alleles described in Mexican Mestizo populations, a set of seven SNP were used for identification of CYP2D6*1, * 2, *3, *4, *5, *10, *17, *29, and other alleles (21,22,26,35,36). TaqMan® probes (Applied Biosystems) were used for this purpose (Table S2). CYP2D6 duplications/multiplications and gene deletion (CYP2D6*5) were evaluated with the copy number variation assay (Applied Biosystems) (Table S2).
Statistical analysis
Population characteristics were described employing category variable frequencies; comparisons were made by using a Chi-squared or Fisher exact test (when needed), at a significance level of P≤0.05. For continuous variables, the mean and median were calculated; comparison between variables was assessed by a Kruskal-Wallis test, with an a posteriori Dunn’s test (when necessary) at a significance level of P≤0.05.
Analyses of SNP allelic and genotypic frequencies, Hardy-Weinberg equilibrium (HWE; Chi-squared test at a significance level of P≤0.05), and linkage disequilibrium (LD) were done with the Arlequin ver. 3.0 software (37). Haplotype frequencies were estimated with the PHASE ver. 2.1 software and the EM algorithm (38,39), and by visual inspection according to the reference haplotype from the Pharmacogenomics Knowledge Base (PharmGKB) (40); diplotypes were conformed as previously suggested (41), and GP inferred with the metabolic activity score (MAS) following the Gaedigk et al. method (4). This method scores each allele in the CYP2D6 diplotype, assigning a value of 0 to null activity alleles, 0.5 to those with decreased activity, and 1.0 to those with normal activity. The score sums of each diplotype are categorized as 0 (gPM); 0.5 (gIM); 1–2 (gNM); and >2 (gUM).
The association between the evaluated TSE and the genetic and non-genetic variables of interest was measured with the two-sided Chi-squared test in univariate models at a P≤0.05 significance level. Comparative parametric and non-parametric tests were applied when needed. Binary logistic regression using dichotomous categories was applied to predict TSE occurrence. Odds ratios (OR), P values, and 95% confidence intervals (95% CI) were calculated. These analyses were run with the SPSS ver. 20 statistical software (SPSS, Inc., Chicago, IL, USA).
Results
Study population
Mean patient age was 50.7 years (range, 31–82 years), with similar proportions of pre- and post-menopausal women (Table 1). Based on Graffar’s socioeconomic scale, 97.2% of participants belonged to classes II–IV, and only 2.8% to class I. Most (77.5%) participants had received chemotherapy prior to initiating tamoxifen treatment, while only 21.1% had exhibited at least one chronic disease. Median length of tamoxifen use was 21 months (range: 3–108 months).
Table 1
| Characteristics | Value |
|---|---|
| Age (years) | 50.7±11.4 |
| BMI (kg/m2) | 29.6±5.2 |
| Age at menarche (years) | 12.3±1.4 |
| Socioeconomic status | |
| Class I | 2 (2.8) |
| Class II | 25 (35.2) |
| Class III | 23 (32.4) |
| Class IV | 21 (29.6) |
| Menopausal status | |
| Pre-menopausal | 36 (50.7) |
| Post-menopausal | 35 (49.3) |
| Chemotherapy before tamoxifen | |
| Yes | 55 (77.5) |
| No | 16 (22.5) |
| At least one other chronic disease | |
| Yes | 15 (21.1) |
| No | 56 (78.9) |
| Contraceptive therapy use | |
| Yes | 26 (36.6) |
| No | 45 (63.4) |
| Time using contraceptive | |
| Therapy (months) | 60 [1–180] |
| Time using TAM (months) | 21 [3–108] |
Data are shown as mean ± SD, number (percentage) or median [range]. BMI, body mass index; SD, standard deviation.
TSEs
Most participants (90.14%) reported at least one TSE. The four most frequent TSE were HF (57.75%), arthralgia (45.07%), headache (43.66%), and cramps (39.44%). All the remaining TSE occurred at a <20% frequency. Moreover, no significant difference in TSE occurrence was identified based on the two tamoxifen treatment length categories (P>0.19) (Table 2). Notably, three patients had been following a 10-year treatment protocol and all had experienced TSE. A relatively small proportion of participants reported severe TSE: 4.23% for HF; 1.41% for arthralgia; 1.41% for headaches; and 1.41% for vomiting (Table S3). No significant difference was found in TSE severity according to length of tamoxifen use (P>0.10; Table S3).
Table 2
| TSE | Tamoxifen length of treatment (months) | P value† | ||
|---|---|---|---|---|
| Overall (n=71) | 3–21 (n=36) | >21 (n=35) | ||
| One TSE, n (%) | 0.43 | |||
| Yes | 64 (90.14) | 31 (86.11) | 33 (94.29) | |
| No | 7 (9.86) | 5 (13.89) | 2 (5.71) | |
| Hot flashes, n (%) | 0.39 | |||
| Yes | 41 (57.75) | 19 (52.78) | 22 (62.86) | |
| No | 30 (42.25) | 17 (47.22) | 13 (37.14) | |
| Arthralgia, n (%) | 0.91 | |||
| Yes | 32 (45.07) | 16 (44.44) | 16 (45.71) | |
| No | 39 (54.93) | 20 (55.56) | 19 (54.29) | |
| Headache, n (%) | 0.19 | |||
| Yes | 31 (43.66) | 13 (36.11) | 18 (51.43) | |
| No | 40 (56.34) | 23 (63.89) | 17 (48.57) | |
| Vomiting, n (%) | 0.43 | |||
| Yes | 7 (9.86) | 5 (13.89) | 2 (5.71) | |
| No | 64 (90.14) | 31 (86.11) | 33 (94.29) | |
| Nausea, n (%) | 0.96 | |||
| Yes | 12 (16.90) | 6 (16.67) | 6 (17.14) | |
| No | 59 (83.10) | 30 (83.33) | 29 (82.86) | |
| Dizziness, n (%) | 0.61 | |||
| Yes | 16 (22.54) | 9 (25.00) | 7 (20.00) | |
| No | 55 (77.46) | 27 (75.00) | 28 (80.00) | |
| Cramps, n (%) | 0.29 | |||
| Yes | 28 (39.44) | 12 (33.33) | 16 (45.71) | |
| No | 43 (60.56) | 24 (66.67) | 19 (54.29) | |
†, Chi-squared test calculated between 3 and 21 and >21 groups. TSE, tamoxifen side effects.
CYP2D6 genotypes/phenotypes
Four SNP corresponded to the HWE (P>0.2), and three were monomorphic (rs59421388, rs35742686, and rs28371706) (Figure S1). Linkage disequilibrium (LD) analysis identified disequilibrium between rs16947 and rs3892097 and rs1065852 (D’=0.99; P<0.05). In addition, disequilibrium was present between rs1065852 and rs3892097 (D’=1.00; P=0.0001). The CYP2D6 haplotype was therefore inferred using only the polymorphic variants (Table 3).
Table 3
| Inferred alleles | Frequencies (%) | Functional status | Diplotypes | Number (%) | MAS | GP (n, %) |
|---|---|---|---|---|---|---|
| *4 | 5.9 | No function | *4J/*5 | 2 (2.8) | 0 | gPM (2, 2.8) |
| *4J | 5.2 | *34/*4J | 3 (4.2) | 1 | gNM (66, 93.0) | |
| *5 | 2.2 | *1/*4 | 6 (8.5) | |||
| *10 | 0.8 | Decreased function | *2/*4 | 3 (4.2) | ||
| *1 | 37.5 | Normal function | *39/*5 | 1 (1.4) |
GP, genetic phenotype; MAS, metabolic activity score; gPM, genetic poor metabolizer; gIM, genetic intermediate metabolizer; gNM, genetic normal metabolizer; gUM, genetic ultra-rapid metabolizer.
Fourteen diplotypes were identified in the population, the most prevalent (32.4%) being CYP2D6*1/*2 (Table 3). Using the MAS method (4) phenotypes were inferred for the four metabolizer categories: gIM (0%); gPM (2.8%); gUM (4.2%); and gNM (93.0%).
TSEs: predictive value of CYP2D6 and other clinical variables
After grouping the cases by their inferred GP no differences were observed in age (P=0.23) or the median number of TSE exhibited per each category (P=0.41) (Table S4); although six patients (9.09%) in the gNM group (n=66) exhibited up to five different TSE. All six studied TSE were reported by participants in the gNM group; arthralgia, vomiting, and cramps were not present in the gUM group; and arthralgia, vomiting, and dizziness were not present in the gPM group. No differences in TSE presence/absence were identified between these three GP (P>0.16) (Table S4).
Uni- and multivariable analyses showed the CYP2D6 GP to be inadequate predictors of the occurrence of at least one TSE (Table S5), and inadequate predictors specifically of HF (Table 4), as well as of the remaining symptoms (data not shown). However, these models did identify chemotherapy prior to tamoxifen initiation as a strong predictor of at least one TSE (Table S5). Chemotherapy and contraceptive use during reproductive age were strong predictors of HF occurrence (Table 4). In the multivariable analysis, chemotherapy was predictive only of headache (Table S6), and pre-menopausal status only of cramps (Table S6).
Table 4
| Explanatory variables | Hot flashes (yes/no) | ||||
|---|---|---|---|---|---|
| Unadjusted | Adjusteda | ||||
| OR (95% CI) | P | OR (95% CI) | P | ||
| Age (years) | |||||
| >40 | Ref. | Ref. | |||
| ≤40 | 0.80 (0.25–2.51) | 0.70 | 0.46 (0.10–2.04) | 0.31 | |
| BMI (Kg/m2) | |||||
| >30 | Ref. | Ref. | |||
| ≤30 | 0.91 (0.34–2.39) | 0.84 | 0.71 (0.22–2.32) | 0.57 | |
| Months using TAM | |||||
| >21 | Ref. | Ref. | |||
| ≤21 | 0.66 (0.26–1.70) | 0.39 | 0.75 (0.24–2.38) | 0.34 | |
| Pre–menopausal | |||||
| No | Ref. | Ref | |||
| Yes | 1.16 (0.44–3.04) | 0.77 | 1.70 (0.44–6.49) | 0.44 | |
| Contraceptive therapy | |||||
| No | Ref. | Ref. | |||
| Yes | 0.22 (0.08–0.61) | 0.003 | 0.21 (0.07–0.67) | 0.008 | |
| Genetic phenotype | |||||
| gNM/gUM | Ref. | Ref. | |||
| gPM | – | – | – | – | |
| Chemotherapy | |||||
| No | Ref. | Ref. | |||
| Yes | 6.17 (1.74–21.83) | 0.003 | 6.98 (1.68–29.09) | 0.008 | |
a, Odds ratio adjusted for all variables listed in table. Ref, reference category; OR, odds ratio; CI 95%, 95% confidence interval; BMI, body mass index; gNM, genetic normal metabolizer; gPM, genetic poor metabolizer; gUM, genetic ultra-rapid metabolizer.
Discussion
One of the main challenges to compliance in adjuvant hormone therapy (e.g., tamoxifen treatment) is the occurrence of TSE during initial treatment stages (14,42). However, this was not observed among the participants, since TSE occurred regardless of the point they were at in the treatment. Indeed, TSE prevalence were higher (P=0.005) in the participants studied here (89%) than in a study of U.S. patients (73%) (15).
Hot flashes (HF) are the most prevalent and widely studied vasomotor symptom (15,43-45), and have been proposed as a suitable marker of therapeutic efficacy (8,43). This agrees with the present findings in which HF were the most prevalent TSE (57.75%). Occurrence did not differ between pre- and post-menopausal women, which do not agree with a study largely attributing HF to pre-menopausal women (15). Only 4.23% of the participants in the present results reported HF as severe, whereas up to 21% were reported as severe in U.S. patients (15).
Some authors have suggested that HF occurrence depends on the adequate endoxifen levels, since this is the most active metabolite produced from tamoxifen biotransformation (8,16), and is associated to CYP2D6 integrity. However, this association has not been found in other studies (9). The present results were more in accordance with in that the CYP2D6 GP were not predictive of any of the studied TSE. By contrast, Goetz et al. (8) reported that gPM patients had lower HF incidence than gNM and the randomized Breast International Group (BIG) 1-98 Trial found that gPM and gIM patients exhibited a higher risk of HF than gNM patients (46). Of note is that two of the patients assigned to the gPM group in the present study lacked TSE predictive value. In another study, SNP suggestive of CYP2D6*41 (related to the gIM phenotype) were associated with fatty liver disease in breast cancer patients under tamoxifen treatment (47).
Since gNM and gUM efficiently generate endoxifen, they would, in theory, be the groups at highest risk of TSE occurrence (48). One study of Italian gUM patients found them to have a higher number of TSE when compared with gPM, gIM and gNM groups (16). Again, the present results exhibited no statistical difference between median TSE numbers among the three identified phenotypes. One study that coincides with the present results is derived from the Adjuvant Tamoxifen and Exemestane in Early Breast Cancer trial and found no association was found between CYP2D6 genotypes/phenotypes and TSE (9). As mentioned above, most previous research in this area has mainly involved populations of Caucasian and Asian origin, leaving geographic areas, such as Latin America acutely underrepresented. How CYP2D6 influences TSE remains unclear and will require extensive further research to determine if it has predictive relevance in clinical practice.
It was noteworthy that tamoxifen use with previous chemotherapy was found to be a strong predictor of HF, an association that remained significant even after adjusting for other variables such as CYP2D6 phenotype. This does not coincide with results from the BIG 1-98 group in which an association was found between HF and CYP2D6 in the group which had received chemotherapy prior to tamoxifen use (46). Indeed, even when taking into account patients who had received chemotherapy (n=55), phenotype was not associated with HF in the present study; although contraceptive use during reproductive age was still a predictive factor (Table S6). Though not a direct contrast, the fact that presence of contraceptive use during reproductive age in the present results was identified as a protective factor against HF development in both the uni- and multivariable models is not supported by a study in the US in which previous use of post-menopausal hormone replacement therapy was reported as a risk factor for development of TSE (15).
Of the alleles responsible for the absence of an association between TSE and CYP2D6 GP found here, two apparent functional variants are worth noting: CYP2D6*34 (13.2%) and *39 (14.7%). The *34 variant was originally reported in 1997 in a European population at a <2.7% frequency (49), and *39 was named a “functional allele” in 2008 after in vitro evaluation (50). Both alleles were also reported in 2010 in a Brazilian population at <1% (51). Samples inferred as CYP2D6*39 could be analyzed with an extended SNP panel including rs1058164, exclusive to CYP2D6*39 (52). Additional analyses could target other variants recently reported in Mexico: rs769258 for CYP2D6*35 (53); rs28735595 for CYP2D6*41 (53); or rs267608 for CYP2D6*53 (21).
The 2.8% frequency for gPM in the present population does not differ (P=0.47) from that reported in a study of women in the US (~5%) (15). In Asians, gPM barely reaches 0.2%, while in Caucasians it is higher (~10%) (54). In another study, CYP2D6*4 was the main allele (70–90% of cases) representing the gPM group (55), but in the present study it was the genotype *4J/*5. To our knowledge this is the first report of a sub-variant form of the *4 allele in Mexican Mestizo patients. The original report for *4J was at a <2% frequency (49), which is lower than the 5.1% observed here. Allele *5 was detected in 2.2% of the present cases. Its frequency varies widely between different populations (2.6–12.5%) (46,56-59), but the present results coincide with those reported for a general population in Mexico (1.3–2.67%) (60).
A possible inherent limitation to the present study is the small sample size of the cross-sectional design, which still had 60% of the power needed to detect significant pharmacogenetic associations (see Methods). This enabled it to identify the less common variants in CYP2D6 (of which only two were classified as gPM). As described in previous paragraphs, the association has been inconsistent between studies. The limited sample size is due to the fact that only current tamoxifen users could be included in the study; this is a result of institutional limitations within the Mexican public health system and the cross-sectional approach. This means no data was collected about patients who could not tolerate this drug, had finished or stopped the treatment with it, or had switched to an aromatase inhibitor. One way to begin addressing this shortfall would be to implement retrospective analyses within the studied population to build a more integrated perspective of tamoxifen efficacy.
Phenotype inference is a complex process that depends heavily on the criteria used. In the present results, for instance, even though no gIM-related alleles (CYP2D6*17, *29, *41 or *59) were identified, the MAS system would have classified them as gNM in the presence of normal function alleles (4). Application of other criteria might have classified them as gIM rather than gNM (41), which could agree with the allele-dosage effect of reduced-function CYP2D6 alleles on reduced plasma endoxifen concentrations (61). The gIM group is therefore operationally defined by separating the gNM/gPM genotype from the gNM (41). Under this scenario the gNM group (93.0%) in the present results would account for 76.1% of the total while the gIM group would be 18.3%. The gIM GP is also very rare in Mexican Mestizos; it has not been reported at all in the country’s southeast (53), and attains only 2.0% frequency in the north (62). Moreover, when the analysis included all four GP (i.e., gPM, gIM, gNM and gUM) they still had no effect on HF prediction (data not shown). Variation in phenotype inference criteria can clearly generate incongruous results in different data sets. Further pharmacokinetic research will help to clarify the causes behind this variation and to better define phenotype groups.
The present findings on side effects in women under tamoxifen treatment are in conspicuous disagreement with some previous research. Generating a broader data foundation that would allow drawing firm conclusions will require a larger sample size that captures the genetic complexity of Mexican Mestizos, and evaluation of the “real” metabolic phenotype.
Conclusions
Hot flashes were the most frequent TSE in the present sample, but CYP2D6 genetic phenotypes were not effective predictors of side effect occurrence or frequency. Chemotherapy prior to tamoxifen treatment and contraceptive use during reproductive age were the only two predictive biomarkers of hot flashes. Allele distribution results showed CYP2D6*34 and *39 to have unusually high frequencies (even after adjusting for the possible presence of other variants reported in other groups from Mexico) in the studied women, which have not been reported in other populations. In conjunction with other reports, the present study contributes to pharmacogenetic characterization of Mexico’s Mestizo populations, which are poorly represented in the literature. Further research is sorely needed to better elucidate the controversial association between CYP2D6 and tamoxifen metabolism.
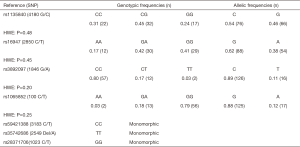
Table S1
| Class | Drugs |
|---|---|
| Inhibitors | |
| Anti-arrhythmic | Amiodarone, propafenone |
| Antibiotics | Quinidine, terbinafine, chloroquine, quinacrine |
| Anticancer drugs | Doxorubicin, lomustine, vinblastine, vincristine, vinorelbine |
| Antihistamines | Chlorphenamine, diphenhydramine, cimetidine, ranitidine |
| Anti-hypertensives | Labetalol, mibefradil |
| Antipsychotics | Chlorpromazine, haloperidol, thioridazine, levomepromazine, fluphenazine |
| Antiretrovirals | Ritonavir, delavirdine |
| Calcium-channel blockers | Diltiazem |
| Inhibitor of monoamine-oxidase | Moclobemide |
| Nonsteroidal anti-inflammatory drugs | Celecoxib |
| Other | Bupropion, metoclopramide, lobeline, yohimbine, encapone |
| Selective serotonine reuptake inhibitors | Citalopram, fluoxetine, paroxetine, fluvoxamine |
| Steroidal anti-inflammatory drugs | Methadone, codeine, dextropropoxyphene |
| Tricyclic antidepressants | Clomipramine, imipramine, desipramine |
| Inductors | |
| Antibiotics | Glucocorticoids, griseofulvin, rifabutin, rifampicin, nafcillin, sulfadimidine |
| Anticonvulsive | Carbamazepine, ethosuximide, phenytoin, primidone, oxcarbazepine, Phenobarbital |
| Antidiabetics | Troglitazone |
| Antigout | Sulfinpyrazone |
| Antiretrovirals | Nelfinavir, nevirapine |
| Hormone replace drugs | Progesterone |
| Nonsteroidal anti-inflammatory drugs | Phenylbutazone, rofecoxib |
| Steroids | Dexamethasone |
Table S2
| SNP | SNP rs | TaqMan probes | CYP2D6 allele targets |
|---|---|---|---|
| 100 C/T | rs1065852 | C__11484460_40 | CYP2D6*4, *10 |
| 1023 C/T | rs28371706 | C__2222771_40 | CYP2D6*17 |
| 4180 G/C | rs1135840 | C__27102414_10 | CYP2D6*2,*4, *10, *17, *29, *39 |
| 2850 C/T | rs16947 | C__27102425_10 | CYP2D6*2, *17, *29, *34 |
| 1846 G/A | rs3892097 | C__27102431_D0 | CYP2D6*4 |
| 2549 Del | rs35742686 | C__32407232_50 | CYP2D6*3 |
| 3183 C/T | rs59421388 | C__34816113_20 | CYP2D6*29 |
| XN/Del | Hs00010001_cn | CYP2D6*XN/CYP2D6*5 |
SNP, single-nucleotide polymorphism; XN, multiplications; Del, deletion.
Table S3
| TSE | Tamoxifen length of treatment (months) | P value† | ||
|---|---|---|---|---|
| Overall (n=71) | 3–21 (n=36) | >21 (n=35) | ||
| Hot flashes, n (%) | 0.77 | |||
| Mild | 28 (39.44) | 16 (44.44) | 12 (34.29) | |
| Moderate | 9 (12.68) | 4 (11.11) | 5 (14.29) | |
| Severe | 3 (4.23) | 2 (5.56) | 1 (2.86) | |
| No | 31 (43.66) | 14 (38.89) | 17 (48.57) | |
| Arthralgia, n (%) | 0.90 | |||
| Mild | 19 (26.76) | 9 (25.00) | 10 (28.57) | |
| Moderate | 12 (16.90) | 7 (19.44) | 5 (14.29) | |
| Severe | 1 (1.41) | 0 (0.00) | 1 (2.86) | |
| No | 39 (54.90) | 20 (55.56) | 19 (54.29) | |
| Headache, n (%) | 0.11 | |||
| Mild | 18 (25.41) | 13 (36.11) | 5 (14.29) | |
| Moderate | 12 (16.93) | 6 (16.67) | 6 (17.14) | |
| Severe | 1 (1.41) | 0 (0.00) | 1 (2.86) | |
| No | 40 (56.34) | 17 (47.22) | 23 (65.71) | |
| Vomiting, n (%) | 0.30 | |||
| Mild | 4 (5.63) | 3 (8.33) | 1 (2.86) | |
| Moderate | 2 (2.82) | 2 (5.56) | 0 (0.00) | |
| Severe | 1 (1.41) | 0 (0.00) | 1 (2.86) | |
| No | 64 (90.14) | 31 (86.11) | 33 (94.29) | |
| Nausea, n (%) | 0.58 | |||
| Mild | 6 (8.45) | 2 (5.56) | 4 (11.43) | |
| Moderate | 6 (8.45) | 4 (11.11) | 2 (5.71) | |
| Severe | 0 (0.00) | 0 (0.00) | 0 (0.00) | |
| No | 59 (83.10) | 30 (83.33) | 29 (82.86) | |
| Dizziness, n (%) | 1.00 | |||
| Mild | 13 (18.31) | 7 (19.44) | 6 (17.14) | |
| Moderate | 3 (4.23) | 2 (5.56) | 1 (2.86) | |
| Severe | 0 (0.00) | 0 (0.00) | 0 (0.00) | |
| No | 55 (77.46) | 27 (75.00) | 28 (80.00) | |
| Cramps, n (%) | 0.29 | |||
| Mild | 14 (19.72) | 8 (22.22) | 6 (17.14) | |
| Moderate | 14 (19.72) | 6 (16.67) | 7 (20.00) | |
| Severe | 0 (0.00) | 0 (0.00) | 0 (0.00) | |
| No | 43 (60.56) | 22 (61.11) | 22 (62.86) | |
†, Fisher’s exact test. TSE, tamoxifen side effects.
Table S4
| TSE | gNM (n=66) | gUM (n=3) | gPM (n=2) | P value |
|---|---|---|---|---|
| Age (years) | 48.50 [31.00–82.00] | 57.00 [48.00–57.00] | 60.00 [55.00–65.00] | 0.23b |
| No. of TSE | 2 [0–5] | 2 [0–3] | 1 [1–1] | 0.41b |
| Hot flashes | 0.63c | |||
| Yes | 37 (56.06) | 2 (66.67) | 2 (100.00) | |
| No | 29 (43.94) | 1 (33.33) | 0 (0.00) | |
| Arthralgia | 0.16c | |||
| Yes | 32 (48.48) | 0 (0.00) | 0 (0.00) | |
| No | 34 (51.52) | 3 (100.00) | 2 (100.00) | |
| Headache | 0.30c | |||
| Yes | 28 (42.42) | 1 (33.33) | 2 (100.00) | |
| No | 38 (57.58) | 2 (66.67) | 0 (0.00) | |
| Vomiting | 1.00c | |||
| Yes | 7 (10.61) | 0 (0.00) | 0 (0.00) | |
| No | 59 (89.39) | 3 (100.00) | 2 (100.00) | |
| Nausea | 0.75c | |||
| Yes | 10 (15.15) | 1 (33.33) | 1 (50.00) | |
| No | 56 (84.85) | 2 (66.67) | 1 (50.00) | |
| Dizziness | 0.73c | |||
| Yes | 15 (22.73) | 1 (33.33) | 0 (0.00) | |
| No | 51 (77.27) | 2 (66.67) | 2 (100.00) | |
| Cramps | 0.38c | |||
| Yes | 27 (40.91) | 0 (0.00) | 1 (50.00) | |
| No | 39 (59.09) | 3 (100.00) | 1 (50.00) |
Data presented as median [range] or number (percentage). b, Kruskal-Wallis test; c, Fisher’s exact test. gNM, genetic normal metabolizer; gPM, genetic poor metabolizer; gUM, genetic ultra-rapid metabolizer; TSE, tamoxifen side effects.
Table S5
| Explanatory variables | One TSE (yes/no) | ||||
|---|---|---|---|---|---|
| Unadjusted | Adjusteda | ||||
| OR (95% CI) | P value | OR (95% CI) | P value | ||
| Age (years) | |||||
| >40 | Ref. | Ref. | |||
| ≤40 | 0.64 (0.11–3.66) | 0.63 | 0.03 (0.001–1.03) | 0.052 | |
| BMI (kg/m2) | |||||
| >30 | Ref. | Ref. | |||
| ≤30 | 0.24 (0.03–2.14) | 0.24 | 0.08 (0.005–1.25) | 0.07 | |
| Months using TAM | |||||
| >21 | Ref. | Ref. | |||
| ≤21 | 0.38 (0.07–2.08) | 0.43 | 0.29 (0.04–2.38) | 0.11 | |
| Pre-menopausal | |||||
| No | Ref. | Ref | |||
| Yes | 2.38 (0.49–11.56) | 0.42 | 12.78 (0.67–244.60) | 0.09 | |
| Contraceptive therapy | |||||
| No | Ref. | Ref. | |||
| Yes | 0.75 (0.15–3.64) | 1.00 | 0.50 (0.07–3.87) | 0.51 | |
| Genetic phenotype | |||||
| gNM/gUM | Ref. | Ref. | |||
| gPM | – | – | – | ||
| Chemotherapy | |||||
| No | Ref. | Ref. | |||
| Yes | 5.78 (1.14–29.30) | 0.04 | 24.28 (1.80–330.26) | 0.02 | |
a, Odds ratio adjusted for all variables listed in table. Ref, reference category; OR, odds ratio; 95% CI, 95% confidence interval; BMI, body mass index; gNM, genetic normal metabolizer; gPM, genetic poor metabolizer; gUM, genetic ultra-rapid metabolizer; TSE, tamoxifen side effects.
Table S6
| Explanatory variables | Headache (yes/no) (n=71) | Cramps (yes/no) (n=71) | Hot flashes (yes/no) (n=55)† | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unadjusted | Adjusteda | Unadjusted | Adjusteda | Unadjusted | Adjusteda | ||||||||||||
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | ||||||
| Age (years) | |||||||||||||||||
| >40 | Ref. | Ref. | Ref. | Ref. | Ref. | Ref. | |||||||||||
| ≤40 | 0.83 (0.26–2.64) | 0.75 | 0.30 (0.07–1.27) | 0.10 | 0.18 (0.04–0.86) | 0.02 | 0.16 (0.03–0.94) | 0.04 | 0.37 (0.11–1.29) | 0.11 | 0.30 (0.06–1.54) | 0.15 | |||||
| BMI (Kg/m2) | |||||||||||||||||
| >30 | Ref. | Ref. | Ref. | Ref. | Ref. | Ref. | |||||||||||
| ≤30 | 0.59 (0.22–1.54) | 0.28 | 0.33 (0.10–1.08) | 0.07 | 0.71 (0.27–1.90) | 0.50 | 0.67 (0.22–2.07) | 0.48 | 0.63 (0.19–2.15) | 0.46 | 0.71 (0.17–2.99) | 0.64 | |||||
| Months using TAM | |||||||||||||||||
| >21 | Ref. | Ref. | Ref. | Ref. | Ref. | Ref. | |||||||||||
| ≤21 | 0.53 (0.21–1.38) | 0.19 | 0.45 (0.14–1.42) | 0.17 | 0.59 (0.23–1.55) | 0.29 | 0.47 (0.15–1.44) | 0.19 | 0.54 (0.17–1.70) | 0.29 | 0.66 (0.17–2.59) | 0.55 | |||||
| Pre–menopausal | |||||||||||||||||
| No | Ref. | Ref | Ref. | Ref | Ref. | Ref. | |||||||||||
| Yes | 2.00 (0.74–5.41) | 0.17 | 2.95 (0.80–10.89) | 0.11 | 0.34 (0.12–0.91) | 0.03 | 0.39 (0.12–1.29) | 0.12 | 0.71 (0.21–2.43) | 0.59 | 1.42 (0.28–7.13) | 0.67 | |||||
| Contraceptive therapy | |||||||||||||||||
| No | Ref. | Ref. | Ref. | Ref. | Ref. | Ref. | |||||||||||
| Yes | 1.17 (0.44–3.10) | 0.75 | 1.90 (0.58–6.24) | 0.29 | 1.20 (0.45–3.23) | 0.71 | 2.37 (0.73–7.78) | 0.15 | 0.12 (0.03–0.42) | 0.0001 | 0.13 (0.04–0.50) | 0.003 | |||||
| Genetic phenotype | |||||||||||||||||
| gNM/gUM | Ref. | Ref. | Ref. | Ref. | Ref. | Ref. | |||||||||||
| gPM | – | – | – | – | 1.56 (0.10–25.93) | 1.00 | 0.89 (0.04–18.61) | 0.94 | – | – | – | – | |||||
| Chemotherapy | |||||||||||||||||
| No | Ref. | Ref. | Ref. | Ref. | NR | NR | |||||||||||
| Yes | 7.80 (1.62–37.65) | 0.004 | 12.33 (2.06–73.65) | 0.006 | 1.58 (0.48–5.17) | 0.45 | 3.13 (0.83–11.83) | 0.09 | |||||||||
a, Odds ratio adjusted for all variables listed in table; †, Calculated in chemotherapy-treated patients. Ref, reference category; OR, odds ratio; CI 95%, 95% confidence interval; BMI, body mass index; gNM, genetic normal metabolizer; gPM, genetic poor metabolizer; gUM, genetic ultra-rapid metabolizer; NR, no required.
Acknowledgments
Thanks are due Dr. Juan Pablo Brito-Miranda, and the oncologists at the UMAE Oncology Department for providing facilities for patient invitations and interviews. Dr. Luis Alfonso Rodríguez-Carvajal provided support with the statistical analyses, and J. Lindsay-Edwards assisted with manuscript editing.
Funding: This research reported here was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.12.27). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures complied with the ethical standards of the IMSS National Clinical Research Ethics Committee (R-2013-785-057), the Dr. Hideyo Noguchi Regional Research Center Review Board for Ethical Research with Human Subjects, and the 1964 Helsinki Declaration and its amendments (as revised in 2013). A written informed consent was obtained individually from all study participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Swaby RF, Sharma C, Jordan V. SERMs for the treatment and prevention of breast cancer. Rev Endocr Metab Disord 2007;8:229-39. [Crossref] [PubMed]
- Desta Z, Ward BA, Soukhova NV, et al. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther 2004;310:1062-75. [Crossref] [PubMed]
-
Pharmacogene Variation Consortium - Gaedigk A, Simon S, Pearce R, et al. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther 2008;83:234-42. [Crossref] [PubMed]
- Hoskins JM, Carey L, McLeod H. CYP2D6 and tamoxifen: DNA matters in breast cancer. Nat Rev Cancer 2009;9:576-86. [Crossref] [PubMed]
- Karle J, Bolbrinker J, Vogl S, et al. Influence of CYP2D6-genotype on tamoxifen efficacy in advanced breast cancer. Breast Cancer Res Treat 2013;139:553-60. [Crossref] [PubMed]
- Saladores P, Mürdter T, Eccles D, et al. Tamoxifen metabolism predicts drug concentrations and outcome in premenopausal patients with early breast cancer. Pharmacogenomics J 2015;15:84-94. [Crossref] [PubMed]
- Goetz MP, Rae J, Suman V, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol 2005;23:9312-8. [Crossref] [PubMed]
- Dezentjé VO, Gelderblom H, Van Schaik RH, et al. CYP2D6 genotype in relation to hot flashes as tamoxifen side effect in a Dutch cohort of the tamoxifen exemestane adjuvant multinational (TEAM) trial. Breast Cancer Res Treat 2014;143:171-9. [Crossref] [PubMed]
- Binkhorst L, Mathijssen RH, Jager A, et al. Individualization of tamoxifen therapy: Much more than just CYP2D6 genotyping. Cancer Treat Rev 2015;41:289-99. [Crossref] [PubMed]
- Early Breast Cancer Trialists’ Collaborative Group. Tamoxifen for early breast cancer: an overview of the randomised trials. Early Breast Cancer Trialists’ Collaborative Group. Lancet 1998;351:1451-67. [Crossref] [PubMed]
- Davies C, Pan H, Godwin J, et al. Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. Lancet 2013;381:805-16. [Crossref] [PubMed]
- Li C, editor. Breast Cancer Epidemiology. In: Principles of Breast Cancer Therapy. New York: Springer; 2010.
- Howell A. Results of the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial after completion of 5 years’ adjuvant treatment for breast cancer. Lancet 2005;365:60-2. [Crossref] [PubMed]
- Lorizio W, Wu AH, Beattie MS, et al. Clinical and biomarker predictors of side effects from tamoxifen. Breast Cancer Res Treat 2012;132:1107-18. [Crossref] [PubMed]
- Rolla R, Vidali M, Meola S, et al. Side Effects Associated with Ultrarapid Cytochrome P450 2D6 Genotype among Women with Early Stage Breast Cancer Treated with Tamoxifen. Clin Lab 2012;58:1211-8. [PubMed]
- Henry NL, Rae JM, Li L, et al. Association between CYP2D6 genotype and tamoxifen-induced hot flashes in a prospective cohort. Breast Cancer Res Treat 2009;117:571-5. [Crossref] [PubMed]
- Goetz MP, Ingle J. CYP2D6 Genotype and Tamoxifen: Considerations for Proper Nonprospective Studies. Clin Pharmacol Ther 2014;96:141-4. [Crossref] [PubMed]
- Luo HR, Poland R, Lin K, et al. Genetic polymorphism of cytochrome P450 2C19 in Mexican Americans: A cross-ethnic comparative study. Clin Pharmacol Ther 2006;80:33-40. [Crossref] [PubMed]
- Dorado P, Sosa-Macias MG, Peñas-LLedó EM, et al. CYP2C9 allele frequency differences between populations of Mexican-Mestizo, Mexican-Tepehuano and Spaniards. Pharmacogenomics J 2011;11:108-12. [Crossref] [PubMed]
- Contreras AV, Monge-Cazares T, Alfaro-Ruiz L, et al. Resequencing, haplotype construction and identification of novel variants of CYP2D6 in Mexican Mestizos. Pharmacogenomics 2011;12:745-56. [Crossref] [PubMed]
- López M, Guerrero J, Jung-Cook H, et al. CYP2D6 genotype and phenotype determination in a Mexican Mestizo population. Eur J Clin Pharmacol 2005;61:749-54. [Crossref] [PubMed]
- Silva-Zolezzi I, Hidalgo-Miranda A, Estrada-Gil J, et al. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc Natl Acad Sci U S A 2009;106:8611-6. [Crossref] [PubMed]
- Rubi-Castellanos R, Martínez-Cortés G, Muñoz-Valle J, et al. Pre-Hispanic Mesoamerican demography approximates the present-day ancestry of Mestizos throughout the territory of Mexico. Am J Phys Anthropol 2009;139:284-94. [Crossref] [PubMed]
- Hertz DL, Rae JM. One step at a time: CYP2D6 guided tamoxifen treatment awaits convincing evidence of clinical validity. Pharmacogenomics 2016;17:823-6. [Crossref] [PubMed]
- Sosa-Macías M, Elizondo G, Flores-Pérez C, et al. CYP2D6 genotype and phenotype in Amerindians of Tepehuano origin and Mestizos of Durango, Mexico. J Clin Pharmacol 2006;46:527-36. [Crossref] [PubMed]
- Power calculator for binary outcome superiority trial. Sealed Envelope Ltd. 2012. 2012. Available online: https://www.sealedenvelope.com/power/binary-superiority/
- Gauderman W, Morrison J. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. 2006. Available online: http://hydra.usc.edu/gxe
- McHutchison JG, Manns M, Patel K, et al. Adherence to combination therapy enhances sustained response in genotype-1-infected patients with chronic hepatitis C. Gastroenterology 2002;123:1061-9. [Crossref] [PubMed]
- Partridge AH, Wang P, Winer E, et al. Nonadherence to adjuvant tamoxifen therapy in women with primary breast cancer. J Clin Oncol 2003;21:602-6. [Crossref] [PubMed]
- Osterberg L, Blaschke T. Adherence to medication. N Engl J Med 2005;353:487-97. [Crossref] [PubMed]
- Andrade SE, Kahler K, Frech F, et al. Methods for evaluation of medication adherence and persistence using automated databases. Pharmacoepidemiol Drug Saf 2006;15:565-74. [Crossref] [PubMed]
- National Cancer Institute: Common Terminology Criteria for Adverse Events (CTCAE) v5.0. Available online: http://ctep.cancer.gov/protocolDevelopment/electronic_applications /ctc.htm
- Graffar M. Une méthode de classification social d’ échantillons de population. Courrier 1956;6:445-9.
- Salazar-Flores J, Torres-Reyes LA, Martínez-Cortés G, et al. Distribution of CYP2D6 and CYP2C19 polymorphisms associated with poor metabolizer phenotype in five Amerindian groups and western Mestizos from Mexico. Genet Test Mol Biomarkers 2012;16:1098-104. [Crossref] [PubMed]
- Lazalde-Ramos BP, Martínez-Fierro Mde L, Galaviz-Hernández C, et al. CYP2D6 gene polymorphisms and predicted phenotypes in eight indigenous groups from northwestern Mexico. Pharmacogenomics 2014;15:339-48. [Crossref] [PubMed]
- Laurent E, Guillaume L, Stefan S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinform Online 2007;1:47-50. [PubMed]
- Stephens M, Smith N, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 2001;68:978-89. [Crossref] [PubMed]
- Stephens M, Scheet P. Accounting for Decay of Linkage Disequilibrium in Haplotype Inference and Missing-Data Imputation. Am J Hum Genet 2005;76:449-62. [Crossref] [PubMed]
- PharmGKB-CYP2D6 haplotype. Available online: https://www.pharmgkb.org/gene/PA128/haplotype
- Hicks JK, Swen J, Thorn C, et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin Pharmacol Ther 2013;93:402-8. [Crossref] [PubMed]
- Rae JM, Sikora M, Henry N, et al. Cytochrome P450 2D6 activity predicts discontinuation of tamoxifen therapy in breast cancer patients. Pharmacogenomics J 2009;9:258-64. [Crossref] [PubMed]
- Baxter SD, Teft W, Choi Y, et al. Tamoxifen-associated hot flash severity is inversely correlated with endoxifen concentration and CYP3A4*22. Breast Cancer Res Treat 2014;145:419-28. [Crossref] [PubMed]
- Gu R, Jia W, Zeng Y, et al. A comparison of survival outcomes and side effects of toremifene or tamoxifen therapy in premenopausal estrogen and progesterone receptor positive breast cancer patients: a retrospective cohort study. BMC Cancer 2012;12:161. [Crossref] [PubMed]
- Chen Y, Dorjgochoo T, Bao P, et al. Menopausal Symptoms among Breast Cancer Patients: A Potential Indicator of Favorable Prognosis. PLoS One 2013;8:e75926. [Crossref] [PubMed]
- Regan MM, Leyland-Jones B, Bouzyk M, et al. CYP2D6 Genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: The breast international group 1-98 trial. J Natl Cancer Inst 2012;104:441-51. [Crossref] [PubMed]
- Wickramage I, Tennekoon K, Ariyaratne M, et al. CYP2D6 polymorphisms may predict occurrence of adverse effects to tamoxifen: a preliminary retrospective study. Breast Cancer (Dove Med Press) 2017;9:111-20. [Crossref] [PubMed]
- Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clin Pharmacol Ther 2011;89:662-73. [Crossref] [PubMed]
- Marez D, Legrand M, Sabbagh N, et al. Polymorphism of the cytochrome P450 CYP2D6 gene in a European population: characterization of 48 mutations and 53 alleles, their frequencies and evolution. Pharmacogenetics 1997;7:193-202. [Crossref] [PubMed]
- Sakuyama K, Sasaki T, Ujiie S, et al. Functional characterization of 17 CYP2D6 allelic variants (CYP2D6.2, 10, 14A-B, 18, 27, 36, 39, 47-51, 53-55, and 57). Drug Metab Dispos 2008;36:2460-7. [Crossref] [PubMed]
- Friedrich DC, Genro JP, Sortica VA, et al. Distribution of CYP2D6 alleles and phenotypes in the Brazilian population. PLoS One 2014;9:e110691. [Crossref] [PubMed]
- Whirl-Carrillo M, McDonagh E, Hebert J, et al. Pharmacogenomics Knowledge for Personalized Medicine. Clin Pharmacol Ther 2012;92:414-7. [Crossref] [PubMed]
- Perez-Paramo YX, Hernandez-Cabrera F, Dorado P, et al. Interethnic relationships of CYP2D6 variants in native and Mestizo populations sharing the same ecosystem. Pharmacogenomics 2015;16:703-12. [Crossref] [PubMed]
- Chin FW, Chan SC, Abdul Rahman S, et al. CYP2D6 Genetic Polymorphisms and Phenotypes in Different Ethnicities of Malaysian Breast Cancer Patients. Breast J 2016;22:54-62. [Crossref] [PubMed]
- Zhou SF. Polymorphism of Human Cytochrome P450 2D6 and Its Clinical Significance. Part I. Clin Pharmacokinet 2009;48:689-723. [Crossref] [PubMed]
- Sistonen J, Fuselli S, Palo JU, et al. Pharmacogenetic variation at CYP2C9, CYP2C19 and CYP2D6 at global and microgeographic scales. Pharmacogenet Genomics 2009;19:170-9. [Crossref] [PubMed]
- Rebsamen MC, Desmeules J, Daali Y, et al. The AmpliChip CYP450 test: cytochrome P450 2D6 genotype assessment and phenotype prediction. Pharmacogenomics J 2009;9:34-41. [Crossref] [PubMed]
- Damodaran SE, Pradhan SC, Umamaheswaran G, et al. Genetic polymorphisms of CYP2D6 increase the risk for recurrence of breast cancer in patients receiving tamoxifen as an adjuvant therapy. Cancer Chemother Pharmacol 2012;70:75-81. [Crossref] [PubMed]
- Hicks JK, Swen JJ, Gaedigk A. Challenges in CYP2D6 phenotype assignment from genotype data: A critical assessment and call for standardization. Curr Drug Metab 2014;15:218-32. [Crossref] [PubMed]
- Cuautle-Rodríguez P, Llerena A, Molina-Guarneros J. Present status and perspective of pharmacogenetics in Mexico. Drug Metabol Drug Interact 2014;29:37-45. [Crossref] [PubMed]
- Mürdter TE, Schroth W, Bacchus-Gerybadze L, et al. Activity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of phase I and II enzymes on their concentration levels in plasma. Clin Pharmacol Ther 2011;89:708-17. [Crossref] [PubMed]
- Alcazar-González GA, Calderón-Garcidueñas A, Garza-Rodríguez M, et al. Comparative study of polymorphism frequencies of the CYP2D6, CYP3A5, CYP2C8 and IL10 genes in Mexican and Spanish women with breast cancer. Pharmacogenomics 2013;14:1583-92. [Crossref] [PubMed]