
An improved understanding of cancer genomics through massively parallel sequencing
Introduction
Cancer is not one disease, but a collection of different diseases that share common features: origination from patients’ own cells and a disrupted regulatory program resulting in uncontrolled growth. Cancers also generally have a genetic origin; changes in the biological blueprint initiate the signaling and regulatory alterations that lead to tumorigenesis. Early work at the beginning of the 20th century suggested that certain chromosomal aberrations could cause unregulated growth in sea urchin eggs, leading to a variety of hypotheses regarding the role of chromosomes in cancer (1). The role of genetic alterations in human cancer was confirmed with the discovery and subsequent classification of the Philadelphia chromosome (2,3). This and other genetic discoveries led to theories of the requirement of multiple genetic events driving clonal selection of tumor cells (4-6). Advances in molecular manipulation played a role in the cloning and identification of the first oncogenes, followed by the discovery of the exact nucleotide change responsible for the oncogenic phenotype (in this case, the genetic changes resulting in the RAS G12V amino acid substitution) (7-9). This body of work has cemented the role of genetic alterations, large and small, in the biology of cancer.
Cancer genes had been discovered prior to the public release of the human genome using techniques such as positional cloning, biological screening assays, and candidate gene studies [for review see (10)]. However, the Human Genome Project has greatly increased our understanding of the structure and contents of our chromosomes (11,12), allowing for the design and execution of experiments that were not previously realistic. In the continued study of cancer genetics, this led to numerous large-scale investigations of many genes across different cancer types, resulting in the identification of a number of new cancer genes. Initial studies focused on smaller groups of genes, including tyrosine kinases (13), a more comprehensive set of kinases (14-16), and other genes (17). Using this approach, BRAF was discovered to contain a common mutation in several cancer types, with a high prevalence in melanoma (18). This discovery eventually resulted in a targeted therapy (vemurafenib) that is commonly used today. Sequencing and sample handling improvements allowed the subsequent investigation of almost all protein coding genes by specific PCR targeting and capillary-based sequencing in 22 breast and colorectal cancers (19), 24 pancreatic cancers (20), and 22 glioblastoma multiforme tumors (21). These improvements were also used to extend kinase sequencing to 210 tumors and matched normal samples, allowing for precise definition and characterization of somatic mutations (22). These studies resulted in better understanding of the specific base change classes that were different across cancer types, passenger vs. driver mutations, and new genes important for cancer biology [such as IDH1 in glioblastoma (21)].
Although previous large-scale genetic studies yielded many new insights into the genetic underpinnings of cancer, there were several limitations. These limitations stemmed from the cost of targeting and sequencing many genes with PCR and capillary sequencing based methods. Few laboratories in the world had the resources to perform such studies, and even those labs had to limit either the number of genes targeted, or the number of tumor samples investigated. And even though sequencing costs had fallen rapidly during the Human Genome Project, costs had not falling rapidly enough to consider sequencing many more samples across all genes, or even whole genomes.
Massively parallel sequencing: technologies
The completion of the Human Genome Project heralded a new vantage point from which to investigate human biology. The National Human Genome Research Institute put forth a vision on the questions and challenges that could now be addressed (23). Part of this document discussed “Quantum Leaps”, or hypothetical technological advances that could “revolutionize biomedical research and clinical practice”. One of these advances focused on improving DNA sequencing technology: “The ability to sequence DNA at costs that are lower by four to five orders of magnitude than the current cost, allowing a human genome to be sequenced for $1,000 or less”. Several new methods [termed massively-parallel sequencing (MPS) or next-generation sequencing] have been developed that essentially increased the numbers of molecules that could be interrogated at the same time [light-based pyrosequencing (24); ligation (25); sequencing-by-synthesis (26); single polymerase (27); patterned nanoarrays (28); semiconductor pH-based pyrosequencing (29)]. Each technology has features and advantages that make each suited for particular applications. Generally, available platforms offer either much longer sequences (suitable for de novo sequencing or other application needing long sequences) or high numbers of sequences (suitable for re-sequencing and variant/mutation detection, which is common in cancer studies). An updated overview of features is listed in Table 1; for a more in depth look at the underlying technology and evaluation of the different platforms, see comparisons from Niedringhaus et al. (30) and Liu et al. (31). Although analysis methods may change slightly depending on the technology, as error models can be different across platforms, many tools address the unique aspects of the more common platforms. The introduction and continuing improvements to these methods have resulted in a cost decrease per whole human genome of about four orders of magnitude (http://www.genome.gov/sequencingcosts/), along with dramatically reduced time to sequence a whole genome. Future technologies, including nanopore-based sequencing, aim to reduce costs and time even further.
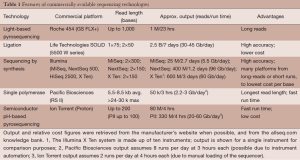
Full table
Technological advances in sequencing have resulted in many new challenges. Many of the challenges result from the hundreds of millions of reads or more that can be generated for a single sample. Even before samples were loaded onto a sequencing instrument, the cost decrease suddenly made PCR-based target selection the main time and cost bottleneck. In response to this challenge, new methods were developed to perform massively parallel target selection, allowing simultaneous targeting the coding regions of hundreds and eventually tens of thousands of genes [Molecular Inversion Probes (32), Microarray-based Genomic Selection (33), and Solution Hybrid Selection (34); compared in (35,36)]. These methods have undergone steady improvement, and form the basis of many commercial products that can target a few genes up to the whole exome. Many commercial offerings are now focusing on cancer with the release of cancer panels for both research and clinical use. More recent improvements to sample preparation are focused on automation, resulting in reduced hands-on time, cost, and variability. Therefore, sequence generation is not only becoming faster and cheaper, it is becoming easier as well.
Massively parallel sequencing: analysis
Even as the cost, time, and human effort required to generate sequence data are all decreasing, the large amount of data is constantly increasing, and is resulting in many analytical challenges. Fundamentally, the output of sequencing instruments has been exceeding the increase in computing power since the introduction of MPS methods (http://www.genome.gov/sequencingcosts/, Moore’s Law is reflected as hypothetical data illustrating the reduction in computing cost). In addition, most computational methods developed for the Human Genome Project were designed with fewer numbers of longer sequences in mind, and therefore were often not able to scale up to analyze tens to hundreds of millions of sequences in short timeframes. Many new computational approaches have been developed at all stages of the data analysis pipeline, allowing for rapid analyses that are tailored to the specific error models and sequence configurations of MPS data. A discussion of the major stages (Figure 1) and some of the more commonly used software tools for DNA sequence analysis follows. For an evaluation of MPS analysis software, see the recent review by Pabinger et al. (37). Analysis of RNA sequence is treated in a final section. Some of the discussed tools have been used in the pan-cancer papers highlighted in the latter part of this review. These studies offer a variety of examples of MPA uses and analysis methods in cancer research.
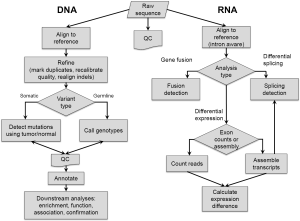
Alignment
The first step in deriving biological meaning from a sequence of bases is determining the sequence location or “alignment” in the human reference genome. The primary challenges in alignment of sequences, or “reads” from MPS experiments include the large number of reads and the shorter length (currently up to ~300 base pairs for high volume technologies). These features result in the need to align many more sequences to the human genome. Each read also has the possibility of many alignments due to the repetitive nature of the human genome and the short length of reads (which may not span repetitive regions and “anchor” the read with unique sequence). The most commonly used methods currently implement the Burrows-Wheeler transform, a lossless data compression technique (38). These include bowtie (39), SOAP2 (http://soap.genomics.org.cn/soapaligner.html) and BWA (40). Other common alignment tools include NovoAlign (http://www.novocraft.com/) and MOSAIK (41).
Genotype determination
Genotype determination (or calling) in cancer is divided into two different types of genetic variation: germline (inherited) variants and somatic mutations, both of which contribute to cancer progression. Inherited variants have been shown to contribute to cancer susceptibility as high-penetrance variants observed in various syndromes (for example TP53, Li-Fraumeni; APC, Familial Adenomatous Polyposis; BRCA1/BRCA2, Hereditary Breast and Ovarian Cancer Syndrome) and as lower penetrance variants contributing to increased risk in a broader portion of the population. Identification of inherited variants is often performed using normal tissue in order to avoid confounding somatic mutations. Current software approaches generally use a Bayesian model, as implemented in SAMtools (42), GATK (43), MPG (35), FreeBayes (44), and others. These software tools can also be used to identify somatic mutations that distinguish tumor cells from normal, but there are caveats to this approach: (I) it is not possible to precisely separate the inherited variants from the somatic mutations without a matched normal sample, even with population genomics data like the 1,000 Genomes project; (II) default settings, especially for filtering, may not be appropriate for cancer somatic mutation data; (III) more powerful approaches exist for directly comparing tumor and matched normal sequences. We have observed (Teer JK, manuscript in preparation) that GATK in particular is tuned for population variation discovery, and the default settings result in very few known somatic mutations passing the Variant Quality Score Recalibration step. Users should carefully examine default settings when attempting to use these methods. Many studies have used these tools to examine germline variants contributing to cancer [for example: (45-47)]. We have also applied exome-wide germline variant discovery to identify cancer-susceptibility variants in a non-cancer cohort of 572 individuals, highlighting some of the expectations and issues that may arise from general genetic screening (48).
To overcome the inability to precisely separate inherited variants from somatic mutations, it has become common practice to sequence both a tumor sample and a matched normal sample from the same individual. Variants observed only in the tumor, and not in the normal sample are inferred to be somatic mutations (tumor specific), and variants observed both in the tumor and in the normal are inferred to be inherited variants (present in all tissues.) Although variants can be determined for tumor and normal samples independently (using the Bayesian tools described above) and then compared, several groups have found that results are improved when directly comparing the aligned sequences of these samples. This approach allows simultaneous examination of all observed bases from both samples at a given position. False positive differences are ignored if there was evidence of a variant in the normal sample (but not enough to actually assign a variant genotype). Additionally, variants at low frequency in the tumor can still be identified if the variant allele frequency is significantly higher compared to the normal. Therefore, a simultaneous consideration of the sequenced bases in both samples increases both sensitivity and specificity, although it does double the experimental cost. This general approach is implemented in VarScan/VarScan 2 (49,50), Strelka (51), SomaticSniper (52), MuTect (53), and Shimmer (54). These programs have been recently used to identify somatic mutations in the various Cancer Genome Atlas (TCGA) mutational landscape studies and others, resulting in a catalog of mutations and frequencies across different tumor types.
Variant annotation
Variant annotation adds context to individual variants, and includes a range of information: whether a variant is coding, the predicted amino acid change, whether that amino acid change might be detrimental to protein function, and whether that amino acid change has been observed before. There are now many tools for gene-based annotation; some can be run locally [ANNOVAR (55), snpEff (56)] while others are run by sending data to external servers [SeattleSeq (57)]. Each tool offers different features and output formats, so users should determine which one (or more) best fits their needs.
In addition to these tools, many databases exist with information about genetic variants (Table 2). These external resources are informative in a variety of different ways. They can help identify how often a variant has been observed in human populations: 1,000 Genomes (58), NHLBI GO Exome Sequencing Project (ESP, http://evs.gs.washington.edu/EVS/), and ClinSeq (59). Resources like the Catalog Of Somatic Mutations In Cancer (COSMIC) (60), TCGA (http://cancergenome.nih.gov/), and the International Cancer Genome Consortium (ICGC) (61) can be used to determine whether a variant has been observed in cancer, in which tumor types, and how frequently. Genotype-phenotype relation datasets can also be used to better understand a variant’s potential impact, and include Online Mendelian Inheritance in Man (OMIM, http://omim.org/), ClinVar (62), the Human Gene Mutation Database (HGMD) (63), and My Cancer Genome (http://www.mycancergenome.org). Finally, Drug Gene Interaction Database (DGIdb) (64) aggregates drug-gene interaction information from a variety of sources to better understand the function and therapeutic relevance of a given gene. These resources can be used to give contextual information to mutations from cancer samples, allowing prioritization of the many mutations using existing knowledge.
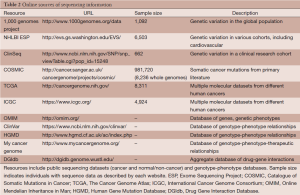
Full table
In silico functional prediction tools use various aspects of the genomes, gene structures, and protein domains to infer the biological impact of a mutation. There are an increasing number of options, including: SIFT (65), SNAP (66), PolyPhen2 (67), and several specific for cancer mutations: CHASM (68), mCluster (69), and transFIC (70). Several tools aggregate the results of other methods to give a meta-score, including Condel (71) and a cancer-specific tool CanPredict (72). FunSeq is specifically designed for detection of functional non-coding mutations, based on evidence of negative selection from the 1,000 Genomes project and functional importance from the ENCODE project (73). The accuracy of these tools varies (74-76), and the general consensus is that they are useful for prioritization, but not for definitive rulings on the effect of a given mutation. Many of the tools demonstrate usage scenarios as part of their published papers, offering readers a chance to evaluate the utility of the resulting information for cancer studies.
Structural variation detection
Larger chromosomal abnormalities have long been known to contribute to cancer development and progression. Massively parallel sequencing experiments can be used to detect chromosomal copy number variants (CNVs), translocations, and other structural variations (SVs). Different approaches are used to detect each type of aberration. CNVs are generally detected using read depth differences. As the read depth in whole genome sequence data is generally homogeneous, deviations from the mean depth can be used to detect CNVs, as in RDXplorer (77) and CNVnator (78). Detecting copy number variation in targeted sequencing experiments using read depth is more challenging, as the genomic capture process introduces significant read depth heterogeneity among regions. Methods to detect somatic CNVs in cancer overcome this issue by directly comparing read depths between a tumor and matched normal. This approach is used by ExomeCNV (79) and VarScan2 (50). Pools of unmatched normal samples are used for comparison by CONTRA (pooled normal control) (80) and EXCAVATOR (single or pooled normals) (81). Finally, two tools use singular value decomposition to normalize each target region across all samples: CoNIFER (82) and XHMM (83).
Although MPS technologies often have shorter read lengths than capillary-based sequencing, paired-end methodologies (in which both ends of a DNA fragment are sequenced) allow inference of the unsequenced part of a molecule. The geometry of the sequence pairs (how far apart from each other they align on the human reference versus the expected fragment size, the orientation with which they align, and the chromosome each pair comes from) allows for indirect detection of structural variation events when the breakpoint lies within the fragment. BreakDancer (84) and SVDetect (85) use this geometry approach to identify read pair orientations indicating a structural anomaly. Other methods use a split-read approach, where the breakpoint can be found in the sequence itself: Pindel (86) and Splitread (87). Packages like DELLY (88) combine short and long insert geometry methods with split-read methods to improve accuracy. Pindel can use BreakDancer results to further refine its detection as well. BreakSeq (89) uses an alternate method: it aligns reads to a custom breakpoint database derived from multiple studies. These methods apply an alternate approach to detect chromosomal rearrangements commonly observed in cancers. Finally, several methods have recently been developed to quantitate the underlying subclonal fractions from paired tumor/normal whole genome sequence based on copy number profiles: somatiCA (90) and THetA (91). Tools like these allow for a better understanding of the heterogeneity of a given tumor, which is likely an important aspect of therapy resistance and relapse.
Significantly mutated
The detection of somatic mutations in different tumor types has made it clear that not all mutations are functional and that many are incidental “passenger” mutations. There is therefore a need to distinguish incidental mutations from those that may be driving the oncogenic process. In silico tools that identified potential drivers based on predicted impact such as CHASM were previously introduced, but the increased number of tumor samples with sequence data allows the use of statistical approaches, which generally identify those positions or genes that are mutated more frequently than expected by chance. This would suggest that these frequently mutated positions might actually be a selection event required for oncogenesis. Tools to perform these calculations include MuSiC (92) and MutSig (93) (SNVs and indels) and Gistic (94) (CNVs). Other methods exist that use alternate non-recurrence approaches (and are therefore complementary to the previous methods). Multi-Dendrix (95) identifies pathways based on gene sets mutated in many patients, but with mutual exclusivity within a patient. Oncodrive-fm (96) identifies potential driver genes based not on mutation recurrence, but on the bias of observed mutations towards higher in silico functional impact. OncodriveCLUST identifies genes with localized mutation clusters (97). ActiveDriver identifies genes of interest based on mutation enrichment in phosphorylation regions (98). These methods are useful tools to distinguish genes important for tumor progression, particularly in cancers with high mutation rates. These tools offer complementary approaches to identify driver genes and mutations, and many have been used in TCGA mutation landscape studies, as well as the pan-cancer studies reviewed below.
RNA-seq
In addition to genetic information gained from sequencing genomic DNA, MPS technologies can be used to quantitate RNA levels. Analysis of RNA sequence data brings several additional challenges. RNA sequence alignment is more difficult due to the absence of introns, which appear as very large gaps when aligning to the complete genome reference. Although methods exist for intron-aware alignment [BLAT (99)], the large number of sequences from a single experiment has required new approaches that scale appropriately. Several new alignment methods have been developed to account for introns and determine accurate alignments across these gaps (including novel exon-exon junctions). These include GSNAP (100), MapSplice (101), SOAPsplice (102), STAR (103), CRAC (104) and Tophat2 (105). Several groups have undertaken comparisons of different aligners, and have shown that while some methods have higher sensitivity and specificity, there is no one method that stands out and all methods have splice-junction misalignments (106-108). Based on the finding of these comparisons, some groups have developed combined approaches that apply a variety of filters to data from existing aligners, thereby improving sensitivity and specificity: RUM (106) and FineSplice (108).
Following alignment, the goal of many RNA sequencing experiments is to identify differentially expressed genes. One class of methods uses approaches similar to those developed for expression microarray analysis: counts across defined regions are compared between groups. These methods have been adapted to address the various differences between array and sequence data. DEseq (109), edgeR (110), and baySeq (111) test for differences assuming a negative binomial distribution of sequence counts at each gene. A recent method, voom (112), allows incorporation of mean-variance relationship estimates into the linear modeling approach implemented in the microarray analysis package limma (113). A second approach generates full transcript models via assembly of sequence reads [Cufflinks (114)], enabling discovery and investigation of known and unknown transcripts. A method for variance estimation and differential expression calculation of these identified transcripts has also been developed [Cuffdiff 2 (115)]. There have been several comparisons of these differential expression techniques (116-118), which reveal various differences between the methods with no one method being optimal under all conditions. Therefore, careful consideration of the pros and cons of each method will be important for data analysis planning.
RNA sequencing also enables identification of gene splicing patterns and quantification of differential isoform expression. Although cutting edge, several methods have been developed to compare relative isoform abundance from RNA-seq data. (Note that there is a larger body of software available to investigate splicing events; the focus here is on differential splicing between samples.) Some methods examine read counts on each exon relative to the whole gene [DEXseq (119)], others consider the exon-exon junction counts representing different isoforms: ALEXA-seq (120), MISO (121), MATS (122), and SpliceSeq (via predefined junction graphs) (123). Tools like SplicingCompass (124) combine these approaches. Others methods merge assemblies or graphs of RNA-seq data into whole or partial isoforms for comparison: Cufflinks (114,115) (whole isoform) and DiffSplice (125) (alternative splicing modules, or informative isoform subregions). To assist with interpretation of complex splicing patterns, many of the tools come with visualization capabilities: DEXseq, ALEXA-seq, MISO (via “sashimi-plots”), SpliceSeq, SplicingCompass, and DiffSplice (via gtf files that can be viewed in genome browsers).
The ability of RNA-seq to determine the exact sequence of an RNA molecule enables the detection of fusion transcripts that result from chromosomal structure variations, including many common gene fusions observed in cancers. Since processed RNA molecules are being interrogated, functionally important structural variants can be detected by examining gene structure. Although the exact breakpoint may not be apparent (if it was in an intron), the functional consequence, including shifts in reading frame, is detectable. Most methods use discordant read pair alignment, split-read alignment, or a combination of both: GSNAP (100), MapSplice (101), FusionMap (126), deFuse (127), TopHat-Fusion (128), ChimeraScan (129), and SOAPfuse (130). Break-Fusion (131) starts with an alignment approach, and then adds local assembly, contig realignment, and a novel chimera score to increase sensitivity for rare events. These methods enable detection of functionally important SVs and oncogenic gene fusions, and can detect variations that targeted genome sequence may not with similar amounts of sequence data.
The landscape of tumor mutations
The key advantage of MPS technologies is the reduced cost and time needed to sequence a sample. This allows for more samples to be investigated than has previously been possible. Although many important genetic mutations have been discovered with earlier methods (see introduction), MPS technologies have enabled the investigation of more samples in a greater variety of cancers. This increase in available information and statistical power has resulted in the identification of many new genes thought to be involved in cancer biology. Efforts by large, international consortia, including TCGA and ICGC, have yielded detailed characterizations of the common somatic genetic alterations in a variety of different tumor types. These findings have been recently reviewed in (132-134).
Putting it all together: pan-cancer studies
Somatic mutations
The advantage of investigating more samples goes beyond better characterization of individual diseases. Datasets generated from individual cancers can now be examined together to compare and contrast across tumor types. A number of groups have recently reported pan-cancer analyses of somatic mutations, resulting in refined lists of likely driver genes. These studies often utilized data from the TCGA project, including ~3,200 samples across 12 different tumors. Tamborero et al. have applied different but complementary approaches for detection of significantly mutated genes to arrive at a high confidence list (135). A total of 291 potential driver genes were identified from the TCGA dataset based on a combination of higher than expected mutation rates, functional mutations, clustered mutations, and mutations in phosphorylation regions. The authors also determined the number of protein altering mutations in each sample, and found that the median driver mutation count per sample is close to previously hypothesized counts (5-7 in epithelial), which suggested that driver detection is “close to saturation”. Although known cancer genes like TP53 and PIK3CA were mutated across tumor types, they also identified 16 genes that were preferentially mutated in one tumor type. Functional bias was specifically used by Reimand et al. to examine mutations in phosphorylation regions (136). Seventy nine genes were mutated at an exact phosphorylation site, and 150 genes were enriched in phosphorylation-related mutations. The authors also identify “network-rewiring mutations” that are predicted to alter kinase-substrate interactions. Both studies demonstrate the importance of including functional considerations when identifying driver genes.
Kandoth et al. used the MuSiC pipeline to identify 127 significantly mutated genes in the TCGA dataset (137). They also showed the distribution of mutations in each tumor type, and found that while most tumor types have mutation rates that fell into a few groups, UCEC and COAD/READ had five and six groups each, some of which had mutation rates >100 fold higher than other groups in each disease. Samples with these high mutation rates were often associated with mutations in DNA repair pathway genes. This observation highlights the fact that tumors of the same type often have very distinct mutational profiles. The authors also detected genes mutated across all and within specific tumor types. Significantly mutated gene pairs were tested for co-occurrence and mutual exclusivity, and many interactions were observed. Mutation status was correlated with clinical outcomes across tumors: associations were found between BAP1, DNMT3A, KDM5C, FBXW7, and TP53 mutations and poor outcome, whereas BRCA2 and IDH1 correlated with improved outcomes. Although not all associations reached statistical significance, this highlighted genes that may be useful prognostic markers across tumor types. Finally, the authors used variant allele frequency to determine clonal heterogeneity, and inferred mutation progression based on the relative mutation allele frequency within a sample.
One of the strengths of the TCGA project is the availability of multiple data types for every sample; the integration of different molecular information sources can yield a deeper knowledge of cancer biology. Ciriello et al. have combined mutation, copy number, and DNA methylation data to identify tumor subclasses (138). They identified “significant functional events”, characterized genes as being altered or not, and identified subclasses based on the alteration signatures. The main finding was the presence of two major groupings based primarily on recurrent point mutations or copy number changes. Interestingly, samples with high numbers of functional events had either many somatic mutations or many copy number changes, not both. The authors described this observation as the “cancer genome hyperbola”. The exception to this finding was TP53, which was highly mutated in copy-change tumors (owing to its role in genome integrity.) The authors found that the subclasses were not unique to individual tumor types, but were variably observed across tumor types. This supports the hypothesis that certain drug combinations could be effective in specific cases across tumor types based on the genomic alteration profile of a tumor. The authors suggested that this type of classification could be used for the design of “basket trials”, where molecular classification, and not tumor type, is used for patient selection.
Using sequence data from the TCGA project and 14 internal projects (for a total of 21 tumor types), Lawrence et al. applied the most recent version of MutSig to identify candidate cancer driver genes (139). Using a larger sample size [4,742], more tumor types, and a more aggressive false discovery rate (0.1), they identified 224 genes using a tumor-type specific analysis, and 114 genes using a combined analysis. The combined analysis yielded an additional 50 genes not seen in the individual analyses, highlighting the benefit of the pan-cancer approach. The individual approach is also still useful, however, as 140 of the 224 genes found to be significant in the individual approach were not significant across all samples (many of these genes were specific to only a few tumor types). The authors then performed a saturation analysis to determine whether sequence information from additional samples would lead to the identification of more significant driver genes. This was accomplished by determining the number of significant genes identified after analyzing smaller and smaller subsets of the existing dataset. They found that the number of significant genes increases approximately linearly, indicating that more genes would be discovered with additional samples. Based on a “restricted hypothesis testing” experiment, they determined that many genes known to be significant in one tumor type would become significant in others, and that adding new tumor types will likely add new “tumor specific” genes. They also suggested that there are still new, infrequently mutated genes to be discovered. A power calculation was employed to determine how many samples would be needed to identify undiscovered genes: they estimated that 650-3,500 samples per tumor type would be needed (more samples needed when a higher mutation rate is observed.)
Recent pan-cancer studies have resulted in an increase in the number of potential cancer driver genes, and better characterization of their mutation frequencies across a variety of tumor types. The main findings from these studies suggest that many significantly mutated genes are commonly mutated across cancer types, supporting the notion of core pathways being important for all cancers. Other genes are more highly mutated in certain tumors, demonstrating that tumor specific pathways are important as well. These overall and tumor specific mutation patterns have proven to be useful to summarize and classify tumors. However, the knowledge of specific mutations in an individual tumor will be critical for understanding the oncogenic progression for that patient’s disease, which will eventually allow for an individualized approach for treatment.
Mutational profiles at the base level
In addition to driver gene discovery, MPS has also been used to summarize the patterns of nucleotide changes within tumors. A recent study has examined mutation profiles in 7,042 cancer samples across 30 tumor types (a combination from TCGA, ICGC, published datasets, and internal datasets) (140). Single nucleotide mutations and their flanking bases were identified, followed by unsupervised clustering methods to define the mutational groups. This resulted in 21 validated mutational signatures. The most prevalent signature (combination of 1A and 1B, seen in 83% of samples) was related to age via possible deamination of 5-methyl-cytosine bases at NpCpG locations. This was the only signature showing a correlation with age of diagnosis. Other signatures were suggested to result from APOBEC-mediated deamination, smoking, UV damage, DNA metabolism changes, and chemotherapy based on the specific mutation profile. In some cases, further associations were performed linking mutation profile to a potential cause (for example, smoking history was positively associated with the presumed smoking signature). A number of signatures resulted from as yet unknown mutagenic events. Combinations of different signatures were observed in most individual cancer samples. The authors also observed “localized substitution hypermutation”, which they previously termed “kataegis” (141). These clusters of C>T and/or C>G mutations were observed in many cancer types in this study, including in >50% of breast, pancreas, lung, CLL, B-cell lymphoma, and ALL samples. The authors suggested that the mutational profile in solid cancers might have resulted from APOBEC deaminase activity. The characterization of these mutational signatures is an important step in understanding the biology of cancer initiation. Knowledge of these signatures will make for useful comparisons against the mutation profiles of a variety of mutagens and other DNA insults and perturbations, especially for the profiles without a known cause. This may eventually lead to a better understanding of molecular processes and environmental agents that cause cancer through genetic mutation.
In the previous study several mutational signatures, including kataegis, were suggested to be the result of increased APOBEC activity (140). Two additional studies have focused on the potential role of the APOBEC deaminases in mutation events leading to tumorigenesis. In the first study, Burns et al. examined APOBEC3B expression levels [previously shown to have a role in breast cancer (142)] across 19 tumor types from the TCGA project (143). They found that APOBEC3B expression is higher in tumor samples (compared to normal) particularly in bladder, head and neck, lung adenocarcinoma, lung squamous carcinoma, and cervical cancer. Mutation counts at CG bases correlated with APOBEC3B expression. APOBEC3B expression levels also correlated with overall mutation counts across tumor types, as well as with clustered mutations (kataegis). In a separate study, Roberts et al. analyzed sequence data from previous publications and the TCGA project (2,680 samples from 14 cancer types) for evidence of APOBEC induced mutations (144). They started by defining a stringent APOBEC motif, and then looked for evidence of enrichment of this APOBEC motif over similar background mutations. They observed clustered mutations, further suggesting that the kataegis process may be related to APOBEC activity. Mutations were examined across tumor types, and a number of tumor types were enriched for APOBEC mutations: bladder, cervical, head and neck, breast, lung adenocarcinoma, and lung squamous carcinoma. From experiments with whole genome data, the authors suggest all cancer types have a background of APOBEC driven mutation, but some cancer types have a heavy enrichment, and can have frequencies as high as 68%. They also showed higher expression of APOBEC family members in tumors, and that this expression (particularly APOBEC3A and APOBEC3B) correlated with number of APOBEC induced mutations.
Massively parallel sequencing has enabled broad investigation and characterization of the patterns of DNA alteration in many different tumor types. In some cases, these patterns agree with known DNA alteration mechanisms. The APOBEC family of deaminases has now been shown to be a major contributor to the mutational burden in cancer. While more patterns are likely to be discovered, there are many today that are not linked to a biological mechanism, suggesting that there is still much to learn about DNA metabolism and the agents that may affect this process.
Beyond nonsynonymous
Although sequencing studies have focused on protein altering mutations, non-coding mutations have been linked to cancer: promoter mutations (145,146) and synonymous mutations (147) in melanoma, and synonymous rare variants in Fanconi Anemia (148). Two recent pan-cancer sequence analyses further reveal the importance of non-coding mutations. Khurana et al. used population genomics approaches to identify functionally important non-coding regions of the human genome using 1,000 Genomes and ENCODE project data (73). Known cancer genes were shown to be the highly enriched for rare coding polymorphisms, suggesting this approach can effectively identify cancer driver genes. Noncoding regions with rare polymorphism enrichment similar to that observed in coding cancer genes (highly constrained regions) were identified and categorized based on known functions. These categories included transcription factor (TF) binding motifs for specific TF families, and were shown to be highly enriched for known human disease variants. Somatic mutations from three cancer types (prostate, breast, medulloblastoma) were identified and characterized based on these constrained categories. The authors found that somatic mutations are enriched for deleterious classes compared to germline variants, including at noncoding positions. Recurrent mutations were observed in noncoding regions, further suggesting noncoding mutations may be driving tumorigenesis. Ninety eight candidate noncoding drivers were identified in breast and prostate cancer genomes using their method implementation, FunSeq.
In a separate approach, Supek et al. investigated the contribution of synonymous (silent) mutations (149). Synonymous mutations in a curated list of potential driver genes were identified in 3,851 samples across 11 tumor types. Synonymous mutations were enriched in oncogenes compared to the matched control genes, but not in tumor suppressor genes (excepting TP53). Interestingly, although genes with frequent non-synonymous mutations also had frequent synonymous mutations, these did not co-occur in the same sample. The authors showed that the most common functional effect of synonymous mutations was the disruption of splicing. Potential splicing disruptions were enriched only in oncogenes, not tumor suppressors. This functional prediction was confirmed using RNA-seq data on 2,000 samples from the same individuals; high association between alternate exon usage and synonymous (but not non-synonymous) mutations was observed. The authors estimate that “half of the putative synonymous drivers are associated with splicing changes”, demonstrating the importance of silent mutations.
These studies demonstrate that the less studied non-coding or synonymous mutations contribute to tumorigenesis. Protein altering mutations are more frequently studied due to the greater understanding of amino acid changes, but the importance of other mutations has now been clearly demonstrated. These studies suggest that samples lacking known driver mutations may have noncoding or synonymous mutations affecting the same genes and pathways. These mutations should therefore be an important target for future studies, as they may account for many cases of “missing drivers” observed in studies focusing exclusively on protein altering mutations.
Non-human genetic contributions to cancer
The majority of sequencing studies have focused on human DNA to identify oncogenic events. However, it is widely appreciated that viruses are significant contributors to human cancers. Viral sequences in tumor cells have been specifically examined in 4,433 samples across 19 tumor types from the TCGA project (150). Viral mRNA was identified by subtraction of sequences that aligned to the human reference genome, characterization against viral reference genomes, and then quantitation across tumor types. Known associations were confirmed: human papillomavirus (HPV) was detected in 96.6% of cervical cancers and hepatitis B virus (HBV) was detected in 32.4% of liver cancers. Interestingly, the authors did not see extensive evidence for Epstein-Barr virus in breast invasive carcinoma, nor cytomegalovirus in glioblastoma multiforme. Although these viruses have been suggested to be involved in these particular cancers, this study revealed no significant role based on detection of viral sequence. They find HPV in other tumors, including head and neck and less commonly in bladder, lung squamous, uterine, and a few colorectal cancers. Viral integrations were observed, and recurrent integrations were seen in or near known cancer genes (MYC, ERBB2, RAD51B, MLL4, FN1). Differential expression among head and neck samples with and without HPV presence was determined: 597 host genes with ≥4-fold expression differences were identified, including cell cycle regulatory genes and oncogenes. Many of these differentially expressed genes had not been previously associated with HPV infection. Application of MPS to examine infectious agents has helped clarify the role viruses play in cancer biology.
Conclusions
MPS has expanded our knowledge of cancer biology due to the greater amount of sequence information that can now be easily generated. This has resulted in the identification of potential cancer driver genes new to specific tumor types, and across all tumor types. Specific mutation patterns and new underlying mutagenesis mechanisms have been clearly defined and provide tools for further identification of mutagenic agents and processes that may contribute to cancer incidence. Functional mutations that do not directly alter protein sequence have been shown, suggesting that many unknown drivers could be explained by non-protein altering mutations in known driver genes. Viral contributions to cancer can now be examined and characterized on a large scale, together with information about the tumor itself. Indeed, genetic information has highlighted molecular similarities across different tumor types, and differences within the same tumor type. The reduced costs of sequencing technologies (as of this writing, $1,000 for a whole human genome) make it possible to further pursue these advances in the broader patient population.
As MPS technologies are recent developments resulting in large amounts of data, analysis methods will continue to mature. Software performance will need to improve due to the continued decrease of sequencing cost (at a rate faster than computational performance is increasing). New methods to integrate the many data sources now becoming available will also be needed, including both for discovery and for annotation with existing information. Many investigators are answering this challenge and more software tools are being developed, therefore standard frameworks for software evaluation will benefit researchers looking to choose from among these new tools. Availability and utility of analysis software is an important component of sequencing research, as it allows any interested investigator to test published findings, and generate their own discoveries.
Recent large-scale sequencing analyses in cancer have described and characterized the mutations observed across a range of tumor types. The current “landscape era” is essential to build our understanding of the molecular processes in cancer, but future research will move beyond description and characterization, and more towards clinical meaning. This future translational work will apply recent knowledge to phenotype and outcome studies; some examples of this have been shown in recent pan-cancer studies. Further research is needed to examine molecular-phenotype associations, including patient survival and drug response, across and within tumor types. An expansion of research in this area will be a critical part of realizing more individualized medicine.
Acknowledgments
The author apologizes to researchers whose work was inadvertently not included in this review.
Funding: This work was supported by the H. Lee Moffitt Cancer Center and Research Institute, but the views expressed are solely the view of the author.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Dung-Tsa Chen and Yian Ann Chen) for the series “Statistical and Bioinformatics Applications in Biomedical Omics Research” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2014.05.05). The series “Statistical and Bioinformatics Applications in Biomedical Omics Research” was commissioned by the editorial office without any funding or sponsorship. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J Cell Sci 2008;121:1-84. [PubMed]
- Nowell PC, Hungerford DA. A Minute Chromosome in Human Chronic Granulocytic Leukemia. Science 1960;142:1497.
- Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 1973;243:290-3. [PubMed]
- Knudson AG Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A 1971;68:820-3. [PubMed]
- Nowell PC. The clonal evolution of tumor cell populations. Science 1976;194:23-8. [PubMed]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759-67. [PubMed]
- Reddy EP, Reynolds RK, Santos E, et al. A point mutation is responsible for the acquisition of transforming properties by the T24 human bladder carcinoma oncogene. Nature 1982;300:149-52. [PubMed]
- Tabin CJ, Bradley SM, Bargmann CI, et al. Mechanism of activation of a human oncogene. Nature 1982;300:143-9. [PubMed]
- Taparowsky E, Suard Y, Fasano O, et al. Activation of the T24 bladder carcinoma transforming gene is linked to a single amino acid change. Nature 1982;300:762-5. [PubMed]
- Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer 2004;4:177-83. [PubMed]
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860-921. [PubMed]
- Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science 2001;291:1304-51. [PubMed]
- Bardelli A, Parsons DW, Silliman N, et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science 2003;300:949. [PubMed]
- Davies H, Hunter C, Smith R, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res 2005;65:7591-5. [PubMed]
- Stephens P, Edkins S, Davies H, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet 2005;37:590-2. [PubMed]
- Bignell G, Smith R, Hunter C, et al. Sequence analysis of the protein kinase gene family in human testicular germ-cell tumors of adolescents and adults. Genes Chromosomes Cancer 2006;45:42-6. [PubMed]
- Wang TL, Rago C, Silliman N, et al. Prevalence of somatic alterations in the colorectal cancer cell genome. Proc Natl Acad Sci U S A 2002;99:3076-80. [PubMed]
- Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949-54. [PubMed]
- Sjöblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science 2006;314:268-74. [PubMed]
- Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008;321:1801-6. [PubMed]
- Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321:1807-12. [PubMed]
- Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature 2007;446:153-8. [PubMed]
- Collins FS, Green ED, Guttmacher AE, et al. A vision for the future of genomics research. Nature 2003;422:835-47. [PubMed]
- Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005;437:376-80. [PubMed]
- Shendure J, Porreca GJ, Reppas NB, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 2005;309:1728-32. [PubMed]
- Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 2008;456:53-9. [PubMed]
- Eid J, Fehr A, Gray J, et al. Real-time DNA sequencing from single polymerase molecules. Science 2009;323:133-8. [PubMed]
- Drmanac R, Sparks AB, Callow MJ, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 2010;327:78-81. [PubMed]
- Rothberg JM, Hinz W, Rearick TM, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 2011;475:348-52. [PubMed]
- Niedringhaus TP, Milanova D, Kerby MB, et al. Landscape of next-generation sequencing technologies. Anal Chem 2011;83:4327-41. [PubMed]
- Liu L, Li Y, Li S, et al. Comparison of next-generation sequencing systems. J Biomed Biotechnol 2012;2012:251364.
- Porreca GJ, Zhang K, Li JB, et al. Multiplex amplification of large sets of human exons. Nat Methods 2007;4:931-6. [PubMed]
- Albert TJ, Molla MN, Muzny DM, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods 2007;4:903-5. [PubMed]
- Okou DT, Steinberg KM, Middle C, et al. Microarray-based genomic selection for high-throughput resequencing. Nat Methods 2007;4:907-9. [PubMed]
- Teer JK, Bonnycastle LL, Chines PS, et al. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res 2010;20:1420-31. [PubMed]
- Clark MJ, Chen R, Lam HY, et al. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol 2011;29:908-14. [PubMed]
- Pabinger S, Dander A, Fischer M, et al. A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform 2014;15:256-78. [PubMed]
- Burrows M, Wheeler DJ. A Block-sorting Lossless Data Compression Algorithm. DEC Systems Research Center: Digital Equipment Corporation, 1994. Available online: http://www.hpl.hp.com/techreports/Compaq-DEC/SRC-RR-124.pdf
- Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [PubMed]
- Lee WP, Stromberg MP, Ward A, et al. MOSAIK: a hash-based algorithm for accurate next-generation sequencing short-read mapping. PLoS One 2014;9:e90581 [PubMed]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078-9. [PubMed]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491-8. [PubMed]
- Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 [q-bio.GN], 2012.
- Walsh T, Lee MK, Casadei S, et al. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci U S A 2010;107:12629-33. [PubMed]
- Chang VY, Basso G, Sakamoto KM, et al. Identification of somatic and germline mutations using whole exome sequencing of congenital acute lymphoblastic leukemia. BMC Cancer 2013;13:55. [PubMed]
- Kanchi KL, Johnson KJ, Lu C, et al. Integrated analysis of germline and somatic variants in ovarian cancer. Nat Commun 2014;5:3156. [PubMed]
- Johnston JJ, Rubinstein WS, Facio FM, et al. Secondary variants in individuals undergoing exome sequencing: screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am J Hum Genet 2012;91:97-108. [PubMed]
- Koboldt DC, Chen K, Wylie T, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 2009;25:2283-5. [PubMed]
- Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568-76. [PubMed]
- Saunders CT, Wong WS, Swamy S, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012;28:1811-7. [PubMed]
- Larson DE, Harris CC, Chen K, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics 2012;28:311-7. [PubMed]
- Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213-9. [PubMed]
- Hansen NF, Gartner JJ, Mei L, et al. Shimmer: detection of genetic alterations in tumors using next-generation sequence data. Bioinformatics 2013;29:1498-503. [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164 [PubMed]
- Cingolani P, Platts A. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80-92. [PubMed]
- Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 2009;461:272-6. [PubMed]
- 1000 Genomes Project Consortium, Abecasis GR, Altshuler D, et al. A map of human genome variation from population-scale sequencing. Nature 2010;467:1061-73.
- Biesecker LG, Mullikin JC, Facio FM, et al. The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome Res 2009;19:1665-74. [PubMed]
- Forbes SA, Bhamra G, Bamford S, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr Protoc Hum Genet 2008;Chapter 10:Unit 10.11.
- International Cancer Genome Consortium. International network of cancer genome projects. Nature 2010;464:993-8. [PubMed]
- Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 2014;42:D980-5. [PubMed]
- Stenson PD, Mort M, Ball EV, et al. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 2014;133:1-9. [PubMed]
- Griffith M, Griffith OL, Coffman AC, et al. DGIdb: mining the druggable genome. Nat Methods 2013;10:1209-10. [PubMed]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 2003;31:3812-4. [PubMed]
- Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res 2007;35:3823-35. [PubMed]
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248-9. [PubMed]
- Carter H, Chen S, Isik L, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res 2009;69:6660-7. [PubMed]
- Yue P, Forrest WF, Kaminker JS, et al. Inferring the functional effects of mutation through clusters of mutations in homologous proteins. Hum Mutat 2010;31:264-71. [PubMed]
- Gonzalez-Perez A, Deu-Pons J, Lopez-Bigas N. Improving the prediction of the functional impact of cancer mutations by baseline tolerance transformation. Genome Med 2012;4:89. [PubMed]
- González-Pérez A, López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet 2011;88:440-9. [PubMed]
- Kaminker JS, Zhang Y, Watanabe C, et al. CanPredict: a computational tool for predicting cancer-associated missense mutations. Nucleic Acids Res 2007;35:W595-8 [PubMed]
- Khurana E, Fu Y, Colonna V, et al. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science 2013;342:1235587 [PubMed]
- Tchernitchko D, Goossens M, Wajcman H. In silico prediction of the deleterious effect of a mutation: proceed with caution in clinical genetics. Clin Chem 2004;50:1974-8. [PubMed]
- Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat 2011;32:358-68. [PubMed]
- Gnad F, Baucom A, Mukhyala K, et al. Assessment of computational methods for predicting the effects of missense mutations in human cancers. BMC Genomics 2013;14:S7. [PubMed]
- Yoon S, Xuan Z, Makarov V, et al. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res 2009;19:1586-92. [PubMed]
- Abyzov A, Urban AE, Snyder M, et al. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 2011;21:974-84. [PubMed]
- Sathirapongsasuti JF, Lee H, Horst BA, et al. Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics 2011;27:2648-54. [PubMed]
- Li J, Lupat R, Amarasinghe KC, et al. CONTRA: copy number analysis for targeted resequencing. Bioinformatics 2012;28:1307-13. [PubMed]
- Magi A, Tattini L, Cifola I, et al. EXCAVATOR: detecting copy number variants from whole-exome sequencing data. Genome Biol 2013;14:R120. [PubMed]
- Krumm N, Sudmant PH, Ko A, et al. Copy number variation detection and genotyping from exome sequence data. Genome Res 2012;22:1525-32. [PubMed]
- Fromer M, Moran JL, Chambert K, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet 2012;91:597-607. [PubMed]
- Chen K, Wallis JW, McLellan MD, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods 2009;6:677-81. [PubMed]
- Zeitouni B, Boeva V, Janoueix-Lerosey I, et al. SVDetect: a tool to identify genomic structural variations from paired-end and mate-pair sequencing data. Bioinformatics 2010;26:1895-6. [PubMed]
- Ye K, Schulz MH, Long Q, et al. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009;25:2865-71. [PubMed]
- Karakoc E, Alkan C, O’Roak BJ, et al. Detection of structural variants and indels within exome data. Nat Methods 2011;9:176-8. [PubMed]
- Rausch T, Zichner T, Schlattl A, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012;28:i333-i339. [PubMed]
- Lam HY, Mu XJ, Stütz AM, et al. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nat Biotechnol 2010;28:47-55. [PubMed]
- Chen M, Gunel M, Zhao H. SomatiCA: identifying, characterizing and quantifying somatic copy number aberrations from cancer genome sequencing data. PLoS One 2013;8:e78143 [PubMed]
- Oesper L, Mahmoody A, Raphael BJ. THetA: inferring intra-tumor heterogeneity from high-throughput DNA sequencing data. Genome Biol 2013;14:R80. [PubMed]
- Dees ND, Zhang Q, Kandoth C, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res 2012;22:1589-98. [PubMed]
- Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214-8. [PubMed]
- Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [PubMed]
- Leiserson MD, Blokh D, Sharan R, et al. Simultaneous identification of multiple driver pathways in cancer. PLoS Comput Biol 2013;9:e1003054 [PubMed]
- Gonzalez-Perez A, Lopez-Bigas N. Functional impact bias reveals cancer drivers. Nucleic Acids Res 2012;40:e169 [PubMed]
- Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics 2013;29:2238-44. [PubMed]
- Reimand J, Bader GD. Systematic analysis of somatic mutations in phosphorylation signaling predicts novel cancer drivers. Mol Syst Biol 2013;9:637. [PubMed]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res 2002;12:656-64. [PubMed]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010;26:873-81. [PubMed]
- Wang K, Singh D, Zeng Z, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res 2010;38:e178 [PubMed]
- Huang S, Zhang J, Li R, et al. SOAPsplice: Genome-Wide ab initio Detection of Splice Junctions from RNA-Seq Data. Front Genet 2011;2:46. [PubMed]
- Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15-21. [PubMed]
- Philippe N, Salson M, Commes T, et al. CRAC: an integrated approach to the analysis of RNA-seq reads. Genome Biol 2013;14:R30. [PubMed]
- Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36. [PubMed]
- Grant GR, Farkas MH, Pizarro AD, et al. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM). Bioinformatics 2011;27:2518-28. [PubMed]
- Engström PG, Steijger T, Sipos B, et al. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat Methods 2013;10:1185-91. [PubMed]
- Gatto A, Torroja-Fungairiño C, Mazzarotto F, et al. FineSplice, enhanced splice junction detection and quantification: a novel pipeline based on the assessment of diverse RNA-Seq alignment solutions. Nucleic Acids Res 2014;42:e71 [PubMed]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [PubMed]
- Hardcastle TJ, Kelly KA. baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 2010;11:422. [PubMed]
- Law CW, Chen Y, Shi W, et al. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014;15:R29. [PubMed]
- Smyth GK. Limma: linear models for microarray data. In: Gentelman R, Carey V, Dudoit R, et al. eds. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005,397-420.
- Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511-5. [PubMed]
- Trapnell C, Hendrickson DG, Sauvageau M, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013;31:46-53. [PubMed]
- Soneson C, Delorenzi M. A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinformatics 2013;14:91. [PubMed]
- Rapaport F, Khanin R, Liang Y, et al. Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol 2013;14:R95. [Epub ahead of print]. [PubMed]
- Seyednasrollah F, Laiho A, Elo LL. Comparison of software packages for detecting differential expression in RNA-seq studies. Brief Bioinform 2013; [PubMed]
- Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res 2012;22:2008-17. [PubMed]
- Griffith M, Griffith OL, Mwenifumbo J, et al. Alternative expression analysis by RNA sequencing. Nat Methods 2010;7:843-7. [PubMed]
- Katz Y, Wang ET, Airoldi EM, et al. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 2010;7:1009-15. [PubMed]
- Shen S, Park JW, Huang J, et al. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res 2012;40:e61 [PubMed]
- Ryan MC, Cleland J, Kim R, et al. SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics 2012;28:2385-7. [PubMed]
- Aschoff M, Hotz-Wagenblatt A, Glatting KH, et al. SplicingCompass: differential splicing detection using RNA-seq data. Bioinformatics 2013;29:1141-8. [PubMed]
- Hu Y, Huang Y, Du Y, et al. DiffSplice: the genome-wide detection of differential splicing events with RNA-seq. Nucleic Acids Res 2013;41:e39 [PubMed]
- Ge H, Liu K, Juan T, et al. FusionMap: detecting fusion genes from next-generation sequencing data at base-pair resolution. Bioinformatics 2011;27:1922-8. [PubMed]
- McPherson A, Hormozdiari F, Zayed A, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol 2011;7:e1001138 [PubMed]
- Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 2011;12:R72. [PubMed]
- Iyer MK, Chinnaiyan AM, Maher CA. ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics 2011;27:2903-4. [PubMed]
- Jia W, Qiu K, He M, et al. SOAPfuse: an algorithm for identifying fusion transcripts from paired-end RNA-Seq data. Genome Biol 2013;14:R12. [Epub ahead of print]. [PubMed]
- Chen K, Wallis JW, Kandoth C, et al. BreakFusion: targeted assembly-based identification of gene fusions in whole transcriptome paired-end sequencing data. Bioinformatics 2012;28:1923-4. [PubMed]
- Collisson EA, Cho RJ, Gray JW. What are we learning from the cancer genome? Nat Rev Clin Oncol 2012;9:621-30. [PubMed]
- Watson IR, Takahashi K, Futreal PA, et al. Emerging patterns of somatic mutations in cancer. Nat Rev Genet 2013;14:703-18. [PubMed]
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell 2013;153:17-37. [PubMed]
- Tamborero D, Gonzalez-Perez A, Perez-Llamas C, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep 2013;3:2650. [PubMed]
- Reimand J, Wagih O, Bader GD. The mutational landscape of phosphorylation signaling in cancer. Sci Rep 2013;3:2651. [PubMed]
- Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333-9. [PubMed]
- Ciriello G, Miller ML, Aksoy BA, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet 2013;45:1127-1133. [PubMed]
- Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014;505:495-501. [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [PubMed]
- Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell 2012;149:979-93. [PubMed]
- Burns MB, Lackey L, Carpenter MA, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 2013;494:366-70. [PubMed]
- Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 2013;45:977-83. [PubMed]
- Roberts SA, Lawrence MS, Klimczak LJ, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 2013;45:970-6. [PubMed]
- Huang FW, Hodis E, Xu MJ, et al. Highly recurrent TERT promoter mutations in human melanoma. Science 2013;339:957-9. [PubMed]
- Horn S, Figl A, Rachakonda PS, et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013;339:959-61. [PubMed]
- Gartner JJ, Parker SC, Prickett TD, et al. Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc Natl Acad Sci U S A 2013;110:13481-6. [PubMed]
- Chandrasekharappa SC, Lach FP, Kimble DC, et al. Massively parallel sequencing, aCGH, and RNA-Seq technologies provide a comprehensive molecular diagnosis of Fanconi anemia. Blood 2013;121:e138-48. [PubMed]
- Supek F, Miñana B, Valcárcel J, et al. Synonymous mutations frequently act as driver mutations in human cancers. Cell 2014;156:1324-35. [PubMed]
- Tang KW, Alaei-Mahabadi B, Samuelsson T, et al. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nat Commun 2013;4:2513. [PubMed]


