
Targeting liquid-liquid phase separation in pancreatic cancer
Introduction
Cancer is composed of uncontrolled growing cells with the potential to disrupt the function of distant organs which is the leading cause of death in the developed world (1). Its incidence is still increasing because more people live to an old age, which is a significant risk factor for cancer (2). Much progress has been achieved and several subtypes of cancer show very a good response to current chemotherapy or immunotherapy. However, the majority of cancer patients suffer extensive therapy but still show very poor prognosis. A significant percentage of cancers such as pancreatic adenocarcinoma shows extremely poor five-year survival that is only about 6% (3). Pancreatic adenocarcinoma is resistant to most of current therapies and only a few effective chemotherapeutic options exist. Identification of novel and effective targets or pathways is important to cure cancer, especially for highly malignant cancers such as pancreatic adenocarcinoma.
For working efficiency, eukaryotic cells have developed a well-controlled compartmental system to spatially organize cellular “workforce” based on functional specialization (4). Membrane-bound organelles are important to achieve such a goal (5). For examples, membrane-bound nuclei are specifically dedicated for DNA and RNA synthesis, while membrane-bounded endoplasmic reticulum and Golgi apparatus are specialized for protein sorting and trafficking. The generation of ATP for energetic needs occurs in membrane-bounded mitochondria. Although membrane-bound organelles are good for high-performance biological processes, these fixed structures have limitations especially when dynamic compartments are needed. In reality, lots of biological processes such as signaling transduction, RNA metabolism, DNA transcription, protein translation as well as chromatin structure are very dynamic and thus membrane-less organelles are more suitable in these cases. It is now generally accepted that membrane-less organelles are formed through liquid-liquid phase separation (LLPS) (6-9). Experimental evidence has indicated that nucleoli, nuclear speckles, paraspeckles, promyelocytic leukaemia (PML) and Cajal bodies within the nuclear envelope, and P-bodies, germ granules and stress granules in cytoplasm are formed through LLPS (10-13). A critical role of LLPS in physiological processes has been recognized, but the molecular mechanism remains elusive. Furthermore, its role in pathologic processes is largely unknown. It is unclear whether LLPS is involved in carcinogenesis and whether abnormal LLPS is a therapeutic target for patients with cancer.
1,6-hexanediol is an aliphatic alcohol that can disrupt weak hydrophobic protein-protein or protein-RNA interactions and thus disrupt LLPS (14,15). However, the biological effect of 1,6-hexanediol on cancer cells and its molecular mechanism are unknown. Interestingly, we found that pancreatic cancer cells are sensitive to 1,6-hexanediol, an inhibitor of LLPS. In a subcutaneous xenograft model, 1,6-Hexanediol inhibits pancreatic cancer growth. RNA-sequencing and reverse transcription polymerase chain reaction (RT-PCR) reveal that 1,6-hexanediol could downregulate MYC transcript levels. Together, we provide proof-of-concept to show that abnormal liquid phase separation might be a novel target for pancreatic cancer as well as other cancers.
Methods
Cell culture-related experiments
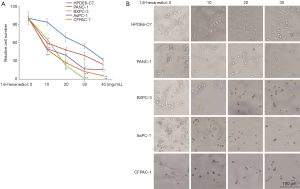
An immortal human pancreatic duct epithelial cell line (HPDE6-C7) and four pancreatic ductal adenocarcinoma (PDAC) cell lines, including BxPC-3, PANC-1, AsPC-1 and CFPAC-1, were purchased from Shanghai Yu Bo Biotech Co., Ltd. All cell lines were maintained in a 5% CO2-humidified incubator at 37 °C. All cells were cultured in DMEM media supplemented with 10% fetal bovine serum, L-glutamine, penicillin, and streptomycin. For CCK8 assay, five cell lines were seeded in 96-well plates (1,000/well) and treated with five different concentrations (0, 10, 20, 30, 40 mg/mL) of 1,6-hexanediol (ALDRICH) for 48 h. Then 20 µL of CCK8 solution was added into each well and the results were examined after 2 hours. In morphologic assay, cells were seeded into 24-well plates and treated by indicated doses of 1,6-Hexanediol for 48 hours. After that, cells were imaged by an optical microscope. All experiments were performed in triplicate.
Atopic BxPC-3 xenograft mouse model in nude mice
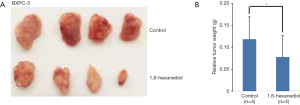
Male nude mice (6–8 weeks of age) housed in the SPF facility of Nantong University were given subcutaneous injection of the same number (1×106/100 µL PBS) of BxPC-3 cells. Mice were randomly divided into a control group (n=4) and a 1,6-hexanediol treatment group (n=4). Animals were treated with water or 1,6-hexanediol (500 mg/kg) by intraperitoneal (i.p.) injection every day for 4 weeks. All animals displayed no obvious health problems during the experimental period. At the end of experiments, all mice were killed. Tumors were resected, weighed, and photographed. All animal studies were approved by Nantong University Animal Care and Use Committee, and handled in strict accordance to institutional protocols.
Quantitative RT-PCR
Total RNA of each sample was extracted using TRIzol reagents (Ambion) as described in the manufacture’s protocol. The quantity of total RNA was determined by Nanodrop 2000. First-strand cDNA library generated from 1 ug of total RNA was performed using Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific). All the mRNA levels were quantified by gene-specific primers using SYBR Green PCR Kit (QIAGEN) on BIO-RAD CFX96 Real-Time System. All reactions were performed in triplicate. The relative expression levels of mRNAs were normalized to GAPDH and calculated by using the comparative threshold cycle method.
RNA-sequencing and analysis
For RNA-sequencing experiments, total RNA was extracted as mentioned above. Ten microliters of total RNA were sent to Genewiz Company for RNA-sequencing. Significantly altered genes with raw reads more than 1,000 in at least one sample were listed. These genes were also analyzed by KEGG (Kyoto Encyclopedia of Genes and Genomes) database. In all cases, the P values of less than 0.05 were considered statistically significant.
Results
Pancreatic cancer cells are sensitive to 1,6-hexanediol
More and more experimental evidence indicates a critical role of protein-mediated LLPS in physiological processes. But the precise role of protein-mediated LLPS in cancer is unknown. We hypothesized that cancer cells might be more sensitive to disruption of LLPS due to many abnormal biological processes in cancer cells. To test our hypothesis, we treated one immortal HPDE6-C7 and four PDAC cell lines, including BxPC-3, PANC-1, AsPC-1 and CFPAC-1, with 1,6-hexanediol (Figure 1A). To our surprise, in a low dose of 1,6-hexanediol (10 mg/mL),almost all control HPDE6-C7 cells survive. However, about 50% of PDAC cells (BxPC-3, PANC-1, AsPC-1 and CFPAC-1) are dead (Figure 1A). With increasing concentrations of 1,6-hexanediol, HPDE6-C7 cells also begin to die (Figure 1A). But in general, compared to control HPDE6-C7 cells, PDAC cells, including BxPC-3, PANC-1, AsPC-1 and CFPAC-1, are more sensitive to 1,6-hexanediol (Figure 1A). To further confirm this result, we monitored the morphologic change of PDAC cells before and after 1,6-hexanediol treatment (Figure 1B). In our tested doses of 1,6-hexanediol, HPDE6-C7 cells show minimal morphologic changes before and after 1,6-hexanediol treatment (Figure 1B). In contrast, all four PDAC cells (BxPC-3, PANC-1, AsPC-1 and CFPAC-1) showed a greater morphologic change even in the lowest dose of 1,6-hexanediol (Figure 1B). Taken together, our results clearly demonstrate that PDAC cells are very sensitive to 1,6-hexanediol, an inhibitor for protein-mediated LLPS.
1,6-hexanediol inhibits pancreatic cancer growth in vivo
Next, we determined whether 1,6-hexanediol could also inhibit pancreatic cancer growth in vivo. Both C57BL/6 and nude mice are well tolerable to 1,6-hexanediol treatment in the dose of 1 g/kg (data not shown). Thus, we select the dose of 500 mg/kg for in vivo experiments. As shown in Figure 2A, 1,6-hexanediol treatment could potently inhibit BxPC-3 pancreatic cancer growth. Statistical analysis further demonstrates that 1,6-hexanediol treatment could significantly limit pancreatic cancer growth in vivo (Figure 2B). Together, we provide convincing evidence to show the inhibitory effect of 1,6-hexanediol on pancreatic cancer both in vitro and in vivo.
1,6-hexanediol downregulates MYC expression
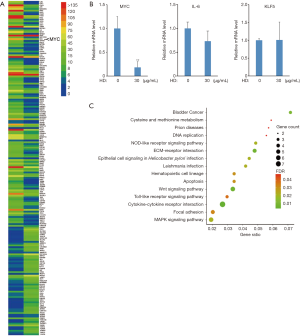
The potent inhibitory effect of 1,6-hexanediol on pancreatic cancer growth encourages us to further understand its molecular mechanism. To globally understand the effect of 1,6-hexanediol on gene transcription, we treated the BxPC-3 cells with 1,6-hexanediol and did RNA-sequencing. Initially, we identified 1,230 genes that are significantly affected by 1,6-hexanediol (data not shown). Among these genes, 189 genes not only have high abundance (raw reads >1,000) at least in one sample (BxPC-3 cells or 1,6-hexanediol-treated BxPC-3 cells) but are also significantly affected by 1,6-hexanediol (Figure 3A). There are 127 significantly downregulated genes and 62 up-regulated genes. Further KEGG pathway analysis of 127 downregulated genes revealed 15 significantly enriched pathways (Figure 3). The most enriched pathways include cytokine-cytokine receptor interaction, WNT signaling pathway, ECM-receptor interaction, MAPK signaling pathway and focal adhesion (Figure 3C). Although there are 62 up-regulated genes, no significant enriched pathway could be identified by KEGG pathway analysis. Interestingly, the most important oncogene MYC is significantly downregulated by 1,6-hexanediol treatment (Figure 3A). To further confirm this finding, we did real-time RT-PCR to examine the expression of MYC before and after 1,6-hexanediol treatment. Consistently, 1,6-hexanediol treatment significantly downregulate MYC but not IL-6 and KLF5 expression (Figure 3B). Our results demonstrate that the LLPS inhibitor, 1,6-hexanediol, could specifically affect several pathways. Most importantly, 1,6-hexanediol could significantly downregulate MYC expression.
Discussion
Based on currently available treatment methods, the majority of cancer patients cannot be cured. Identification of novel targets is important for developing an efficient therapeutic regimen for cancer patients. The abnormal protein-mediated LLPS has been linked to neurodegenerative diseases such as Alzheimer’s disease (16,17). It was recently shown that the Alzheimer’s disease-associated proteins such as tau and FUS, have the ability to form LLPS, and abnormal LLPS of these proteins might be the cause of neurodegenerative diseases (18,19). However, the pathological role of LLPS in other diseases remains enigmatic. Here, we provide proof-of-concept that cancer cells might have abnormal protein-mediated LLPS and cancers cells are more sensitive to disruption of LLPS. However, the proteins that are involved in abnormal LLPS in cancer cells are still undetermined in the current study.
1,6-hexanediol is an aliphatic alcohol that can disrupt LLPS and thus affect the function of membrane-less organelles (14,15). But how does it affect gene transcription globally is unknown. We did RNA-sequencing to examine the change of gene expression before and after 1,6-hexanediol treatment. The most affected genes that are enriched in pathways include cytokine-cytokine receptor interaction, WNT signaling pathway, ECM-receptor interaction, MAPK signaling pathway and focal adhesion (Figure 3B). Several cytokines, such as IL-1B, CSF3, CXCL1, IL-8 and IL-1A, are significantly downregulated by 1,6-hexanediol. These significantly downregulated cytokines are recently shown to play roles in pancreatic cancer development. IL-1A and IL-1B signaling through IL-1R promotes tumor fibrosis and chemoresistance in pancreatic cancer (20,21). Pancreatic cancer cells produce high amounts of G-CSF to recruit neutrophil-like cells for maintaining high immunosuppressive microenvironment (22). Interactions between IL-8 or CXCL1 with their receptors, CXCR1 and CXCR2, sustain pancreatic cancer progression (23,24). All these important cytokines are significantly downregulated by 1,6-hexanediol. Thus, we believed that 1,6-hexanediol might significantly affect tumor microenvironment. The underlying molecular mechanism about how 1,6-hexanediol attenuates the expression of these cytokines has not been solved in this study.
One of the most important downregulated genes upon 1,6-hexanediol treatment is MYC. MYC is a transcription factor that contributes to multiple cancer development (25,26). It becomes an extensively studied therapeutic target in cancer (27). But inhibition of the function of MYC is really a challenging task and there are still no widely accepted methods. It is interesting that 1,6-hexanediol could significantly downregulate MYC expression. Although KRAS, CDKN2A, TP53, and SMAD4 are four main genetic lesions that contribute to the failure of therapies, deciphering vulnerabilities of these genetic lesions remains challenging (28). MYC is considered as a non-redundant signaling hubs under several important signaling pathways and essential for maintaining PDAC (29). The significant downregulation of MYC might contribute to the inhibitory function of 1,6-hexanediol to pancreatic cancer. In the majority of cancer, MYC is over-expressed and, in some cases, due to activation of super-enhancers. Recently, protein-mediated LLPS separation has been hypothesized to play an important role in the organization and function of super-enhancers. Thus, 1,6-hexanediol might disrupt the function of super-enhancers via inhibition of LLPS, but it still needs experimental data to verify this hypothesis. It seems that bromodomain-containing protein-4 (BRD4) inhibitors or super-enhancer inhibitors can downregulate MYC expression. However, the detailed molecular mechanism regarding how 1,6-hexanediol reduces MYC expression is still undetermined. One potential mechanism is that 1,6-hexanediol disrupts LLPS and subsequently disrupts the function of MYC-super enhancers. However, the relationship between LLPS and super-enhancers is still largely unknown.
In summary, we provide proof-of-concept that besides neurodegenerative diseases, abnormal LLPS might be involved in pancreatic cancer. Pancreatic cancer cells might be more addictive to LLPS to support their malignancy. Disruption of LLPS might be a valuable way to treat pancreatic cancer. But global inhibition of LLPS, such as by 1,6-hexanediol, also has toxicity to normal cells. Identification of the proteins involved in abnormal LLPS in pancreatic cancer will allow us to develop more specific methods to treat pancreatic cancer.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.01.06). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This article does not contain any studies with human participants or animals performed by any of the authors. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics CA Cancer J Clin 2012;65:87-108. [Crossref] [PubMed]
- Pawelec G, Derhovanessian E, Larbi A. Immunosenescence and cancer. Crit Rev Oncol Hematol 2010;75:165-72. [Crossref] [PubMed]
- Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med 2014;371:1039-49. [Crossref] [PubMed]
- Aguilera-Gomez A, Rabouille C. Membrane-bound organelles versus membrane-less compartments and their control of anabolic pathways in Drosophila. Dev Biol 2017;428:310-7. [Crossref] [PubMed]
- Blank HM, Totten JM, Polymenis M. CDK control of membrane-bound organelle homeostasis. Cell Cycle 2006;5:486-8. [Crossref] [PubMed]
- Darling AL, Liu Y, Oldfield CJ, et al. Intrinsically Disordered Proteome of Human Membrane-Less Organelles. Proteomics 2018;18:e1700193. [Crossref] [PubMed]
- Uversky VN. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol 2017;44:18-30. [Crossref] [PubMed]
- Uversky VN. Protein intrinsic disorder-based liquid-liquid phase transitions in biological systems: Complex coacervates and membrane-less organelles. Adv Colloid Interface Sci 2017;239:97-114. [Crossref] [PubMed]
- Mitrea DM, Kriwacki RW. Phase separation in biology; functional organization of a higher order. Cell Commun Signal 2016;14:1. [Crossref] [PubMed]
- Brangwynne CP, Eckmann CR, Courson DS, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009;324:1729-32. [Crossref] [PubMed]
- Brangwynne CP. Phase transitions and size scaling of membrane-less organelles. J Cell Biol 2013;203:875-81. [Crossref] [PubMed]
- Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A 2011;108:4334-9. [Crossref] [PubMed]
- Nott TJ, Petsalaki E, Farber P, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 2015;57:936-47. [Crossref] [PubMed]
- Shulga N, Goldfarb DS. Binding dynamics of structural nucleoporins govern nuclear pore complex permeability and may mediate channel gating. Mol Cell Biol 2003;23:534-42. [Crossref] [PubMed]
- Kroschwald S, Maharana S, Mateju D, et al. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife 2015;4:e06807. [Crossref] [PubMed]
- Banani SF, Lee HO, Hyman AA, et al. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 2017;18:285-98. [Crossref] [PubMed]
- Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol 2014;30:39-58. [Crossref] [PubMed]
- Patel A, Lee HO, Jawerth L, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 2015;162:1066-77. [Crossref] [PubMed]
- Ambadipudi S, Biernat J, Riedel D, et al. Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat Commun 2017;8:275. [Crossref] [PubMed]
- Zhang D, Li L, Jiang H, et al. Tumor-Stroma IL1beta-IRAK4 Feedforward Circuitry Drives Tumor Fibrosis, Chemoresistance, and Poor Prognosis in Pancreatic Cancer. Cancer Res 2018;78:1700-12. [Crossref] [PubMed]
- Zhuang Z, Ju HQ, Aguilar M, et al. IL1 Receptor Antagonist Inhibits Pancreatic Cancer Growth by Abrogating NF-kappaB Activation. Clin Cancer Res 2016;22:1432-44. [Crossref] [PubMed]
- Pickup MW, Owens P, Gorska AE, et al. Development of Aggressive Pancreatic Ductal Adenocarcinomas Depends on Granulocyte Colony Stimulating Factor Secretion in Carcinoma Cells. Cancer Immunol Res 2017;5:718-29. [Crossref] [PubMed]
- Fu S, Chen X, Lin HJ, et al. Inhibition of interleukin 8/CX-C chemokine receptor 1,/2 signaling reduces malignant features in human pancreatic cancer cells. Int J Oncol 2018;53:349-57. [PubMed]
- Seifert L, Werba G, Tiwari S, et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature 2016;532:245-9. [Crossref] [PubMed]
- Wolpaw AJ, Dang CV. MYC-induced metabolic stress and tumorigenesis. Biochim Biophys Acta Rev Cancer 2018;1870:43-50. [Crossref] [PubMed]
- Kress TR, Sabo A, Amati B. MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer 2015;15:593-607. [Crossref] [PubMed]
- Whitfield JR, Beaulieu ME, Soucek L. Strategies to Inhibit Myc and Their Clinical Applicability. Front Cell Dev Biol 2017;5:10. [Crossref] [PubMed]
- Wirth M, Mahboobi S, Kramer OH, et al. Concepts to Target MYC in Pancreatic Cancer. Mol Cancer Ther 2016;15:1792-8. [Crossref] [PubMed]
- Hessmann E, Schneider G, Ellenrieder V, et al. MYC in pancreatic cancer: novel mechanistic insights and their translation into therapeutic strategies. Oncogene 2016;35:1609-18. [Crossref] [PubMed]


