Characterization of microRNA expression profiles by deep sequencing of small RNA libraries in leukemia patients from Naxi ethnic
Introduction
MicroRNAs (miRNAs), a class of small noncoding RNAs, regulate the expression of other genes by targeting protein-coding transcripts in the post-transcriptional regulation (1). Approximately, 1,500 miRNAs were annotated in the miRNA database (miRBase) (2) and up to 60% of protein-coding genes were estimated to be regulated by miRNAs (3). miRNAs interact with 3'UTR of messenger RNAs (mRNAs) through base pairing and lead to their degradation, destabilization, or repression of translation through RNA-induced silencing complex (RISC), a complex of multiple proteins and miRNA-mRNA adduct (4,5). Occasionally, these miRNAs can also up-regulate gene expression (6).
It has been reported that miRNAs are essential for normal mammalian development and are involved in many biological processes, such as cell proliferation, differentiation, apoptosis, and metabolism, and their involvement in cancer has sparked increased interest in miRNA biology (7-9). Convincing evidence has indicated that miRNAs are key regulators of hematopoiesis (10,11). Leukemia is a group of hematopoietic neoplasms featuring impaired hematopoiesis and bone marrow failure caused by clonal expansion of undifferentiated myeloid precursors (12). The disease is frequently found to have recurrent chromosomal aberrations and gene mutations, and mostly carry driver mutations relevant to patient clinical outcomes (13). It is grouped by how quickly the disease develops (acute or chronic) as well as by the type of blood cell that is affected (lymphocytes or myelocytes). There are four main types of leukemia, including acute lymphocytic leukemia (ALL), chronic lymphocytic leukemia (CLL), acute myelocytic leukemia (AML), and chronic myelocytic leukemia (CML).
Recently, most researchers use next-generation sequencing (NGS) to profile miRNAs and then identify novel miRNAs associated with a disease of interest (14,15). However, most cohort studies were focused on the major populations such as Asia, America, Europe people etc. Little were known about the molecular profile changes (DNA mutations, RNA transcript and miRNAs, etc.) in the Chinese minorities. The Naxi people are one of the unique ethnic groups in Yunnan and live in a high concentration. In the present study, we analyzed differential expression profiles of miRNAs by quantitative small RNA NGS in six leukemia patients and one normal control. All these people are from Naxi ethnic minorities located in the northwest of Yunnan province, and considered to be an area of low incidence. This approach facilitated in identifying several miRNAs relevant to different leukemia pathogenesis. Additionally, we also verified six miRNAs by quantitative real time-polymerase chain reaction (qRT-PCR) in human leukemic cell lines.
Methods
Patient cohort and sample collection
With informed consent and ethics approval, six leukemia patients (case 2 and case 3 have AML; case 4, case 5, and case 7 have ALL; case 6 has CML) and one healthy person (case 1) were recruited from Tumor Hospital in Yunnan Province. All seven person are from Nakhi (Naxi) ethnic minorities, which considered to be an area of low incidence. The samples were collected in 1.5 mL microcentrifuge tubes, immediately snap-frozen in liquid nitrogen and stored at −80 °C until further processing.
RNA isolation and deep sequencing
Total RNA isolation was carried out from peripheral blood or cell lines using TRIzol® Reagent (Invitrogen) according to the manufacturer’s instructions. The RNA concentration was measured by Nanodrop ND-100 (Thermo Scientific, Waltham, USA), and RNA quality was assessed by the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, USA). Small RNA sequencing libraries were constructed following IlluminaHTruSeqTM Small RNA Sample Preparation protocol. In brief, 3' and 5' adapters were ligated to sRNA population and ligated RNAs were purified by running on urea-PAGE. Modified sRNAs were reverse transcribed to generate cDNA libraries and PCR amplified to add unique index sequence to each library.
Sequencing data analysis
The sequences were converted into FASTQ format and de-multiplexing were performed using Illumina bcl2fastq2 software version 2.17 (http://support.illumina.com/downloads/). Adaptor trimming was performed using the FASTQ Toolkit App of Illumina BaseSpace (http://basespace.illumina.com/apps/). Quality of the sequenced reads before and after adapter trimming was evaluated using FastQC software (http//www.bioinformatics.babraham.ac.uk/projects/fastqc/). Cleaned sequences were aligned to the most recent mirBASE database release 21 (http://mirBase.org/) using the Small RNA App of Illumina BaseSpace. Quantification of miRNA expression was performed by counting aligned reads to miRNA genes.
Novel miRNA prediction
The novel miRNA prediction strategy is based on first removing all known RNAs including those derived from exonic regions and then identifying those that are derived from intronic and intergenic regions. Identifying novel miRNAs from sequencing data commonly apply a combination of (I) evaluating miRNA secondary structures; and (II) ranking miRNA candidates by utilizing existing annotation or evolutionary conservation of the mature miRNA sequence. Structure threshold parameters of most algorithms are often optimized based on animal miRNA precursor structures.
Prediction of miRNA targets
The most probable targets of the differentially regulated miRNAs were predicted by two criteria. (I) Prediction by established target prediction software, which facilitate in identifying all the target genes using intersection between miRanda and miTarget; (II) inverse correlation in expression pattern between miRNA and coding genes. This approach compares the expression of putative genes with the different regulated miRNAs, and only genes that show inverse correlation to the miRNA levels were considered as most putative targets of the select miRNA.
qRT-PCR
qRT-PCR was performed to determine expression of six miRNAs: hsa-miR-181b-5p, hsa-miR-181a-3p, hsa-miR-181a-5p, hsa-miR-342-3p, hsa-miR-450a-5p, hsa-miR-1255a in three different human leukemic cell lines (Jurkat, HL-60, K-562). cDNA synthesis was performed using M-MLV Reverse Transcriptase (Promega) according to the manufacturer’s protocol. All samples were run in three replicates. Ct values for miRNAs were normalized against U6 and the relative expression was calculated using 2−ΔCt (Gene-U6) method. All the RT-qPCR primers are provided by RIBOBIO (http://www.ribobio.com/), the item number was listed in Table S1.
Statistical analysis
All bioinformatics-associated statistical analyses were performed using the R package for statistical computing. One-way analysis of variance (ANOVA) was calculated and analyzed using GraphPad Prism. P<0.05 is considered as statistically significant.
Results
Small RNAs sequencing and annotation
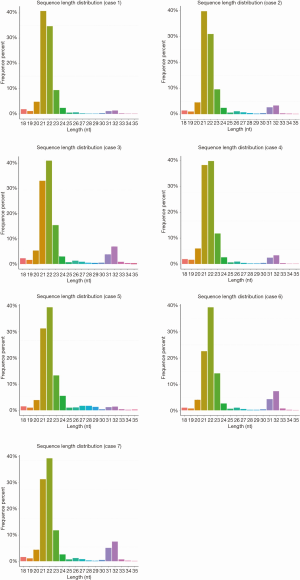
sRNAs from six leukemia patients (case 2 to case 7) and 1 normal control (case 1) were used and size selected by gel electrophoresis, then sequenced using a Solexa platform (Illumina, San Diego, USA). First, we removed the adaptor sequences from the sequence reads, and only those reads that were greater than 10 nucleotides were considered for further analysis. On average, ~13 million reads mapping to the human genome were obtained per sample (Figure 1A). The length of the detected sequences varied between 18 and 32 nucleotides in all these samples (Figure S1), similar to some other studies in animals (18–35 nucleotides) and plants (18–30 nucleotides). Then, we clustered these sequences on the basis of sequence similarity and performed similarity searches using specific databases (miRNAs, rRNA, tRNA, snRNA, snoRNAs). The frequency of reads mapping to miRNAs ranged from 59% to 71% and had a median of 65% (Figure 1B). In total, 1,392 known miRNAs (miRBase 21) were detected in at least one of seven samples sequenced (Table 1). To detect the proportion of mRNA derived by degradation, we checked if there are some sequences derived from intronic and exonic regions, and found that the median read frequency is 7.572% (Figure 1C). Additionally, a small fraction of reads mapped to rRNA, tRNA, snRNA, snoRNA, together comprising a frequency of ~0.8% (Figure 1C).
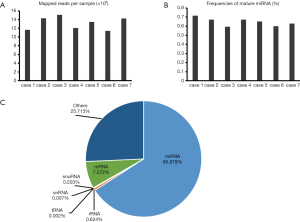
Table 1
| Types | Total | Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 |
|---|---|---|---|---|---|---|---|---|
| Mapped mature miRNA | 1,392 | 942 | 894 | 1,021 | 909 | 981 | 938 | 918 |
| Mapped pre-miRNA | 1,156 | 828 | 793 | 895 | 830 | 857 | 836 | 820 |
miRNA, microRNA.
Identification of candidate novel miRNA genes
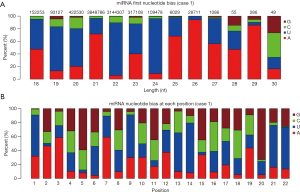
The representative hairpin structure of miRNA precursor is normally used to identify putative novel miRNAs. In this study, we integrated two miRNA prediction software, miRNAEVo (16) and mirdeep2 (17) for the new miRNA prediction. As shown in Table 2. Ninety-nine mature miRNAs, 26-star miRNAs, and 102 miRNA precursors were detected in at least one of the seven samples sequenced. Almost the same number of candidate novel miRNAs was obtained from each sample. During miRNA processing, Dicer enzyme plays an important role from precursor miRNAs to mature miRNAs. Due to the specificity of the enzyme cleavage sites, the first base of miRNAs might display bias. So, we then analyzed the first base distribution of miRNAs in different length, and the base distribution of all miRNAs. As observed, the nucleotide distribution of 18–30 nucleotides novel miRNAs at the first nucleotide position had a strong bias (Figures 2,S2). Additionally, the base distributions of all miRNAs display that there are bias at each position (Figures 2,S3).
Table 2
| Types | Total | Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 |
|---|---|---|---|---|---|---|---|---|
| Mapped mature miRNA | 99 | 53 | 53 | 63 | 62 | 45 | 63 | 57 |
| Mapped star miRNA | 26 | 10 | 8 | 14 | 9 | 10 | 8 | 8 |
| Mapped pre-miRNA | 102 | 57 | 57 | 71 | 64 | 49 | 65 | 59 |
miRNA, microRNA.
miRNA expression pattern
In order to identify miRNAs relevant to different types of leukemia, we divided our samples into four groups, acute leukemia (jxzl), chronic leukemia (mxzl), myeloid leukemia (sxzl) and lymphocytic leukemia (lxzl). To compare the expression levels between different samples, it is necessary to normalize against the read count by calculating a normalization factor. Here, the absolute sequence reads were transformed into transcript abundance by normalizing the data in “transcripts per million (TPM)” for each library first. The expression levels ranged from less than 10 to more than 100,000 counts (Figure S4). Thus, the sequencing data revealed a wide range of expression levels spanning five orders of magnitude.
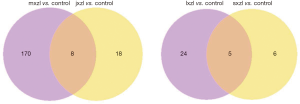
Using an arbitrary P value <0.05 and two-fold change cut-off, we found that in comparison to control, 21 and 125 miRNAs were up-regulated in jxzl and mxzl respectively, while 5 and 53 miRNAs were down-regulated in jxzl and mxzl respectively (Figure 3; Tables S2-S7). Venn diagram displayed that 18 miRNAs specifically regulated in jxzl and 170 miRNAs in mxzl (Figure S5). We then compared the differently expressed miRNAs between jxzl and mxzl, and found that 120 miRNAs were up-regulated and 203 miRNAs were down-regulated in jxzl. The up-regulated genes include six novel miRNAs (novel_581, novel_133, novel_692, novel_224, novel_401, novel_94) and several known miRNAs, such as hsa-miR-103a-2-5p, hsa-miR-320b, hsa-miR-320c, hsa-miR-1180-3p, hsa-let-7a-5p, hsa-let-7b-5p, hsa-let-7c-5p, and hsa-let-7d-5p. And the downregulated genes include novel_498, novel_377, novel_580, hsa-miR-582-5p, hsa-miR-582-3p, has-miR-450b-5p, and has-miR-450a-5p (Tables S2-S7). These differentially expressed miRNAs might play crucial role in regulating the rate of the disease development.
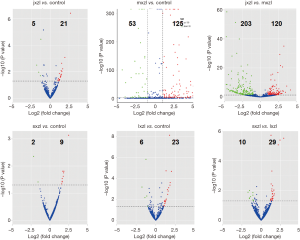
Next, we compared the differently expressed miRNAs between the sxzl or lxzl with control. As shown, 9 and 23 miRNAs were up-regulated in sxzl and lxzl, 2 and 6 miRNAs were down-regulated in sxzl and lxzl (Figure 3; Tables S2-S7). When compared the differently expressed miRNAs in sxzl and lxzl, the results showed that 29 known miRNAs were up-regulated and 10 known miRNAs were downregulated, including hsa-miR-181-5p, hsa-miR-181a-3p, hsa-miR-181b-5p, hsa-miR-342-3p, hsa-let-7f-1-3p, and hsa-miR-30a-5p (Tables S2-S7). These miRNAs might play an important role in the specific type of blood cell development.
Predicted targets of differentially regulated miRNAs
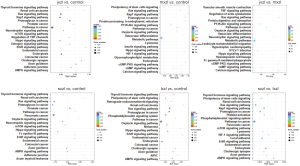
In order to understand the biological role of the differentially regulated miRNAs, it is necessary to figure out the putative targets of the detected miRNAs. In this study, two approaches were employed for target prediction. First, predict target using two popular software tools, miRanda, and miTarget. Then, compare the expression levels between miRNA and putative target genes. Those genes that coexist in two databases, and show inversion correlation to its miRNAs were considered to be the targets. KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis was performed for predicted targets. We analyzed the pathways of differently expressed genes between jxzl and control, and found that the most significant gene-enrichment included “pathways in cancer”, “Rap1 signaling pathways”, “Ras signaling pathways”, and “endocytosis”. However, when compared between mxzl and control, the most significant gene-enrichment was metabolic pathways. Similar results were found in jxzl and mxzl, which suggests that the metabolic pathway is responsible for the development of leukemia. Then we compared the pathways of differently expressed genes between sxzl and control, between lxzl and control (Figure 4). The results display that cancer development is the most significant pathway, which indicates that the miRNAs play an important role in determining the type of cancer and its development.
Validation of candidate miRNAs by qRT-PCR
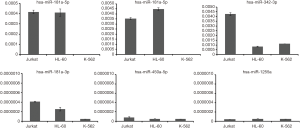
To validate these findings, we examined the expression of six miRNAs, hsa-miR-181b-5p, hsa-miR-181a-3p, hsa-miR-181a-5p, hsa-miR-342-3p, hsa-miR-450a-5p, and hsa-miR-1255a in three different human leukemic cell lines (Jurkat, HL-60, K-562) by RT-qPCR (Figure 5). As known, Jurkat is acute lymphoid leukemia, HL-60 is AML, and K-562 is CML and consistent with the small RNAseq results, we found that hsa-miR-181a-5p and hsa-miR-181b-5p displayed higher expression in Jurkat and HL-60 than that in K-562. However, the expression level of hsa-miR-342a-3p in Jurkat was higher than other two (HL-60 and K-562). And hsa-miR-181a-3p showed different expression in three cell lines (Jurkat had highest and K-562 had lowest). Additionally, both hsa-miR-450a-5p and hsa-miR-1255a displayed low expression in all three cell lines. These results suggest that hsa-miR-181b-5p, hsa-miR-181a-3p, hsa-miR-181a-5p, and hsa-miR-342-3p showed different expression patterns in different types of cancer cells, and hsa-miR-450a-5p, hsa-miR-1255a were dysregulated in all the leukemia cells.
Discussion
The study of miRNA expression has a certain guiding role in clinical practice as the abnormal expression of miRNA may be used as a prognostic indicator for leukemia patients (18,19). However, this is based on a large number of races and there was very little research on ethnic minorities. To provide accurate treatment for minority people, the differences between Han people in genetic characteristics and molecular maps need to be analyzed. In this study, 6 patients from the Naxi ethnic minority were selected for miRNA analysis, the Naxi ethnic group lives in the western Yunnan Plateau in the northwest of Yunnan Province, with unique national culture and regional characteristics. This is the first report of the Naxi ethnic miRNA expression. In spite of the small number of examples, it has laid a certain foundation for the follow-up study.
In this study, six patients from the Naxi ethnic minority were selected for miRNA analysis, the Naxi ethnic group located in the western Yunnan plateau, the northwest of Yunnan province, with a unique national culture and regional characteristics. Our paper, for the first time, studied the miRNA expression in Naxi people. Although the number of samples is limited, it lays the foundation for future research.
Certain investigations indicated that some miRNAs can affect the directional differentiation of hematopoietic cells, and associated with the pathogenesis of leukemia and lymphoma. Understanding and clarifying the relationship between miRNAs and leukemia cells can help to reveal the molecular mechanism of leukemia, and miRNAs and specific target genes can also be used to achieve targeted therapeutics. Till now, several miRNAs expressed tags, as well as miRNAs, responsible for the AML and CLL have been reported (18,20,21). For example, it was found that frequent deletions and down-regulation of miRNA15 and miRNA16 were happened in ALL (22); Garzon et al. found that the expression of miR-191 and miR-199a was up-regulated in AML, and its high expression was associated with poor prognosis in AML patients (18). Dixon-McIver et al. also confirmed that t(15;17) chromosome heterotopia is related to the upregulation of miR-127, miR-154, miR-299, miR-323, miR-368 and miR-370 (19).
In the present study, deep sequencing technology was used to quantify miRNA expression in six leukemia patients from Naxi ethnic. From these samples, we observed several interesting findings, including the discovery of novel miRNAs and a valuable list of differentially expressed miRNAs in chronic and acute leukemia, as well as lymphocytic and myelocytic leukemia. Our major findings are a list of expressed miRNAs (1,392 known and 125 candidate novels) in leukocytes and the discovery that the differential expression of miRNAs is most abundant in the pathway of cancer. These 125 candidate miRNAs should be further studied to exclude the false positive results and find the specific miRNAs after more samples would be collected. Our qRT-PCR results show convincingly that such miRNAs are likely to be real and not a result of an artifact of sequencing.
The known miRNAs found to be enriched in acute leukemia were members of the let-7 family, miR185 and miR320, which were also reported in peripheral blood mononuclear cells using “Taqman miRNA assay” (23). Therefore, our results are in agreement with previous studies based on a different methodology. There are multiple mechanisms that are likely to regulate miRNA levels similar to that of mRNAs.
Additionally, we found for the first time that novel-94, 692, 581, 580, 498, 401, 377, 224 and 133 showed abnormal expression in leukemia patients. We also validate our RNAseq data by RT-qPCR in three leukemia cell lines, the expression of hsa-miR-181b-5p, hsa-miR-181a-3p, hsa-miR-181a-5p, hsa-miR-342-3p, hsa-miR-450a-5p, and hsa-miR-1255a were consistent with our sequence data. Most interestingly, WT1 gene was predicted to be the target of miR-1255a by TargetScan v7.1. As known, WT1 was highly expressed in peripheral blood or narrow bone of most AML patients, and supposed to be the marker of detecting minimal residual disease (MRD). Our results indicated that the expression of miR-1255a was significantly down-regulated in AML patients, which suggested that the WT1 gene might be regulated by miR-1255a. This new finding provides a new direction for the further research.
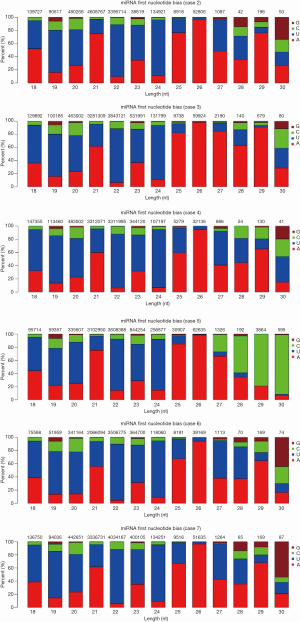
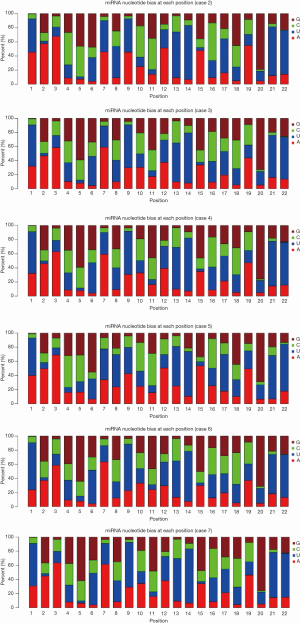
Table S1
| Species | Gene name | Item number |
|---|---|---|
| Human | hsa-miR-181b-5p | MIMAT0000257 |
| Human | hsa-miR-181a-3p | MIMAT0000270 |
| Human | hsa-miR-181a-5p | MIMAT0000256 |
| Human | hsa-miR-342-3p | MIMAT0000753 |
| Human | hsa-miR-450a-5p | MIMAT0001545 |
| Human | hsa-miR-1255a | MIMAT0005906 |
Table S2
| sRNA | jxzl readcount | Control readcount | Log2 fold change | P value | Q value |
|---|---|---|---|---|---|
| Up-regulated | |||||
| hsa-miR-1255a | 55.92721169 | 3.347829955 | 2.7099 | 3.84E-07 | 0.000219 |
| hsa-miR-99b-3p | 27.37110049 | 2.231886637 | 1.9569 | 0.000831 | 0.094769 |
| hsa-miR-1270 | 26.99996407 | 6.69565991 | 1.5303 | 0.002276 | 0.21624 |
| hsa-miR-34a-5p | 177.0151767 | 11.15943318 | 1.6723 | 0.005981 | 0.34091 |
| hsa-miR-335-3p | 24.84632076 | 5.579716592 | 1.4625 | 0.00816 | 0.42281 |
| hsa-miR-139-5p | 226.9695179 | 26.78263964 | 1.5594 | 0.009461 | 0.44938 |
| hsa-miR-1226-3p | 29.98544425 | 10.04348987 | 1.2247 | 0.013456 | 0.5463 |
| hsa-miR-99b-5p | 298.7417542 | 31.24641292 | 1.4743 | 0.015335 | 0.5463 |
| hsa-miR-149-5p | 10.96456363 | 0 | 1.4556 | 0.017281 | 0.56172 |
| hsa-miR-8485 | 11.14732578 | 0 | 1.4116 | 0.020799 | 0.56172 |
| hsa-miR-3117-3p | 15.64362549 | 0 | 1.4008 | 0.021681 | 0.56172 |
| hsa-miR-7854-3p | 6.27480693 | 0 | 1.3922 | 0.022724 | 0.56315 |
| hsa-miR-3656 | 7.953821644 | 1.115943318 | 1.3472 | 0.02591 | 0.61537 |
| hsa-miR-652-5p | 18.29332169 | 6.69565991 | 1.101 | 0.032129 | 0.67828 |
| hsa-miR-23b-5p | 11.61892026 | 3.347829955 | 1.1754 | 0.037711 | 0.72538 |
| hsa-miR-4433a-3p | 188.5453394 | 20.08697973 | 1.2659 | 0.03874 | 0.72538 |
| hsa-miR-4433b-5p | 188.154882 | 20.08697973 | 1.2614 | 0.03945 | 0.72538 |
| hsa-miR-1271-5p | 21.83756653 | 8.927546547 | 1.0088 | 0.042338 | 0.75415 |
| hsa-miR-6786-3p | 5.197110923 | 0 | 1.2277 | 0.043122 | NA |
| hsa-miR-495-3p | 9.17037747 | 0 | 1.2207 | 0.04373 | 0.75534 |
| hsa-let-7e-5p | 245.6468219 | 77.00008897 | 1.1127 | 0.048766 | 0.81754 |
| Down-regulated | |||||
| hsa-miR-450a-5p | 147.230475 | 369.3772384 | −1.2423 | 3.55E-05 | 0.005056 |
| hsa-miR-642a-5p | 1.521240321 | 12.2753765 | −1.795 | 0.003171 | NA |
| hsa-miR-642b-3p | 1.521240321 | 12.2753765 | −1.795 | 0.003171 | NA |
| hsa-miR-3690 | 3.130164922 | 30.1304696 | −1.5273 | 0.010778 | 0.47255 |
| hsa-miR-4435 | 5.723576538 | 20.08697973 | −1.3048 | 0.019909 | 0.56172 |
Table S3
| sRNA | yyp | lfz | Log2 fold change | P value | Q value (Storey et al. 2003) |
|---|---|---|---|---|---|
| Up-regulated | |||||
| hsa-miR-143-3p | 11,510.45 | 241.2097 | 5.5765 | 0 | 0 |
| hsa-miR-146b-5p | 2,419.833 | 410.3383 | 2.56 | 0 | 0 |
| hsa-miR-148a-3p | 70,326.8 | 17,813.54 | 1.9811 | 0 | 0 |
| hsa-miR-199b-5p | 4,105.741 | 19.46322 | 7.7207 | 0 | 0 |
| hsa-miR-20a-5p | 4,805.136 | 1,288.331 | 1.8991 | 0 | 0 |
| hsa-miR-21-5p | 28,309.18 | 4,955.872 | 2.5141 | 0 | 0 |
| hsa-miR-221-3p | 1,682.975 | 202.9544 | 3.0518 | 0 | 0 |
| hsa-miR-222-3p | 1,349.408 | 157.4507 | 3.0994 | 0 | 0 |
| hsa-miR-223-3p | 12,870.55 | 343.7607 | 5.2265 | 0 | 0 |
| hsa-miR-23a-3p | 3,295.188 | 544.0304 | 2.5986 | 0 | 0 |
| hsa-miR-24-3p | 2,006.894 | 377.5864 | 2.4101 | 0 | 0 |
| hsa-miR-26a-5p | 30,229.04 | 10,386.38 | 1.5412 | 0 | 0 |
| hsa-miR-27a-3p | 1,451.578 | 245.2365 | 2.5654 | 0 | 0 |
| hsa-miR-27b-3p | 2,315.865 | 444.9694 | 2.3798 | 0 | 0 |
| hsa-miR-3074-5p | 1,987.784 | 376.6468 | 2.3999 | 0 | 0 |
| hsa-miR-30b-5p | 1,378.356 | 171.0078 | 3.0108 | 0 | 0 |
| hsa-miR-30c-5p | 3,641.622 | 1,384.439 | 1.3953 | 0 | 0 |
| hsa-miR-582-3p | 1,384.41 | 6.711454 | 7.6884 | 0 | 0 |
| hsa-miR-181a-5p | 3,281.849 | 1,623.366 | 1.0155 | 1.48E-303 | 3.76E-302 |
| hsa-miR-142-3p | 1,687.895 | 415.5732 | 2.0221 | 6.37E-297 | 1.57E-295 |
| hsa-miR-340-5p | 1,226.519 | 188.5919 | 2.7012 | 1.57E-272 | 3.75E-271 |
| hsa-miR-145-5p | 815.2829 | 14.7652 | 5.787 | 2.17E-236 | 4.87E-235 |
| hsa-miR-199a-3p | 769.2115 | 37.31568 | 4.3655 | 8.56E-220 | 1.82E-218 |
| hsa-miR-146a-5p | 925.4946 | 168.0548 | 2.4613 | 2.56E-192 | 5.17E-191 |
| hsa-miR-155-5p | 773.0902 | 86.84621 | 3.1541 | 3.57E-191 | 7.03E-190 |
| hsa-miR-29a-3p | 752.4669 | 121.0746 | 2.6357 | 5.12E-165 | 9.83E-164 |
| hsa-miR-361-3p | 1,385.262 | 605.5074 | 1.1939 | 2.54E-150 | 4.75E-149 |
| hsa-miR-28-3p | 612.7393 | 79.06093 | 2.9542 | 8.02E-146 | 1.43E-144 |
| hsa-miR-221-5p | 503.0006 | 29.79886 | 4.0772 | 1.77E-141 | 2.90E-140 |
| hsa-miR-374b-5p | 1,284.321 | 598.7959 | 1.1009 | 2.90E-129 | 4.48E-128 |
| hsa-miR-374c-3p | 1,283.281 | 598.3932 | 1.1007 | 3.87E-129 | 5.85E-128 |
| hsa-miR-197-3p | 506.0279 | 64.42996 | 2.9734 | 2.81E-121 | 3.95E-120 |
| hsa-miR-106b-5p | 830.0408 | 296.512 | 1.4851 | 2.32E-111 | 3.20E-110 |
| hsa-miR-450a-5p | 392.8835 | 44.42982 | 3.1445 | 6.05E-98 | 7.80E-97 |
| hsa-miR-582-5p | 341.3252 | 1.610749 | 7.7273 | 1.88E-87 | 2.24E-86 |
| hsa-miR-223-5p | 404.8034 | 76.91326 | 2.3959 | 1.84E-83 | 2.16E-82 |
| hsa-miR-181a-3p | 254.5749 | 6.442996 | 5.3042 | 8.11E-76 | 9.39E-75 |
| hsa-miR-181b-5p | 262.1431 | 28.72502 | 3.19 | 1.55E-66 | 1.72E-65 |
| hsa-miR-424-5p | 206.9899 | 8.053745 | 4.6838 | 1.89E-61 | 2.06E-60 |
| hsa-miR-769-5p | 203.5842 | 21.47665 | 3.2448 | 1.37E-52 | 1.46E-51 |
| hsa-miR-342-3p | 218.2476 | 30.06731 | 2.8597 | 4.92E-52 | 5.10E-51 |
| hsa-miR-450b-5p | 163.7566 | 6.308767 | 4.6981 | 5.14E-49 | 5.25E-48 |
| hsa-miR-618 | 171.514 | 16.91286 | 3.3421 | 2.58E-45 | 2.60E-44 |
| hsa-miR-100-5p | 148.9041 | 4.563789 | 5.028 | 4.66E-45 | 4.64E-44 |
| hsa-miR-140-5p | 211.8146 | 40.40295 | 2.3903 | 1.87E-44 | 1.84E-43 |
| hsa-miR-27a-5p | 131.4972 | 7.651057 | 4.1032 | 2.03E-38 | 1.94E-37 |
| hsa-miR-424-3p | 187.0288 | 40.67141 | 2.2012 | 5.36E-37 | 5.08E-36 |
| hsa-miR-148a-5p | 121.6586 | 12.75176 | 3.2541 | 3.56E-32 | 3.34E-31 |
| hsa-miR-19b-3p | 255.8993 | 104.296 | 1.2949 | 1.96E-31 | 1.82E-30 |
| hsa-miR-150-5p | 292.6996 | 138.6586 | 1.0779 | 3.01E-30 | 2.76E-29 |
| hsa-miR-425-3p | 221.5587 | 84.69855 | 1.3873 | 3.45E-29 | 3.12E-28 |
| hsa-miR-651-5p | 116.6447 | 19.32899 | 2.5933 | 8.89E-27 | 7.77E-26 |
| hsa-miR-542-3p | 87.88556 | 4.832247 | 4.1849 | 2.15E-26 | 1.84E-25 |
| hsa-miR-99b-5p | 81.0742 | 3.758414 | 4.431 | 7.61E-25 | 6.37E-24 |
| hsa-miR-24-2-5p | 92.89949 | 11.27524 | 3.0425 | 5.09E-24 | 4.17E-23 |
| hsa-miR-10a-5p | 112.6714 | 23.0874 | 2.2869 | 1.24E-23 | 1.01E-22 |
| hsa-miR-181d-5p | 69.53271 | 1.073833 | 6.0169 | 1.33E-21 | 1.04E-20 |
| hsa-miR-145-3p | 68.39748 | 2.281894 | 4.9056 | 1.47E-21 | 1.13E-20 |
| hsa-miR-4521 | 61.86992 | 3.355727 | 4.2045 | 4.42E-19 | 3.25E-18 |
| hsa-miR-361-5p | 92.52108 | 20.67128 | 2.1622 | 7.27E-19 | 5.30E-18 |
| hsa-miR-345-5p | 120.0504 | 42.9533 | 1.4828 | 1.60E-17 | 1.15E-16 |
| hsa-miR-3065-3p | 57.61282 | 4.161101 | 3.7914 | 2.95E-17 | 2.11E-16 |
| hsa-miR-338-5p | 57.32901 | 4.161101 | 3.7842 | 3.62E-17 | 2.57E-16 |
| hsa-miR-660-5p | 156.8507 | 75.16828 | 1.0612 | 1.01E-16 | 7.08E-16 |
| hsa-miR-126-5p | 121.3748 | 51.54397 | 1.2356 | 4.02E-15 | 2.73E-14 |
| hsa-miR-181c-3p | 48.7202 | 0 | 6.9187 | 7.60E-15 | 5.07E-14 |
| hsa-miR-362-5p | 80.22278 | 22.1478 | 1.8568 | 1.09E-14 | 7.20E-14 |
| hsa-miR-199a-5p | 59.03185 | 10.73833 | 2.4587 | 8.23E-14 | 5.40E-13 |
| hsa-miR-125a-5p | 49.38242 | 5.503392 | 3.1656 | 8.57E-14 | 5.57E-13 |
| hsa-miR-152-3p | 56.856 | 10.06718 | 2.4977 | 1.67E-13 | 1.07E-12 |
| hsa-miR-34c-5p | 40.30059 | 1.073833 | 5.23 | 2.27E-13 | 1.44E-12 |
| hsa-miR-16-2-3p | 94.98074 | 40.40295 | 1.2332 | 3.88E-12 | 2.42E-11 |
| hsa-miR-9-5p | 45.78753 | 7.24837 | 2.6592 | 1.25E-11 | 7.73E-11 |
| hsa-miR-505-3p | 60.7347 | 17.04709 | 1.833 | 2.30E-11 | 1.42E-10 |
| hsa-miR-28-5p | 40.30059 | 4.966476 | 3.0205 | 3.09E-11 | 1.89E-10 |
| hsa-miR-652-5p | 30.36735 | 0 | 5.2367 | 1.96E-10 | 1.18E-09 |
| hsa-miR-4286 | 29.61053 | 0 | 4.9779 | 3.41E-10 | 2.03E-09 |
| hsa-miR-143-5p | 26.67786 | 0 | 5.7376 | 2.80E-09 | 1.65E-08 |
| hsa-miR-146b-3p | 55.53157 | 19.46322 | 1.5126 | 4.95E-09 | 2.88E-08 |
| hsa-miR-34a-5p | 25.73183 | 1.342291 | 4.2608 | 8.02E-09 | 4.64E-08 |
| hsa-miR-21-3p | 29.89434 | 5.234934 | 2.5136 | 8.36E-08 | 4.71E-07 |
| hsa-miR-330-3p | 27.52928 | 4.026872 | 2.7732 | 9.65E-08 | 5.39E-07 |
| hsa-miR-574-3p | 53.16651 | 23.0874 | 1.2034 | 2.85E-07 | 1.57E-06 |
| hsa-miR-874-3p | 19.77189 | 0 | 4.6176 | 3.30E-07 | 1.80E-06 |
| hsa-miR-29c-3p | 31.69178 | 8.456432 | 1.906 | 8.89E-07 | 4.76E-06 |
| hsa-let-7e-5p | 32.25939 | 9.261806 | 1.8004 | 1.35E-06 | 7.16E-06 |
| hsa-miR-335-3p | 17.40683 | 0 | 4.6969 | 1.61E-06 | 8.43E-06 |
| hsa-miR-374b-3p | 22.32615 | 4.42956 | 2.3335 | 6.88E-06 | 3.45E-05 |
| hsa-miR-365a-3p | 22.89376 | 5.637621 | 2.0218 | 1.81E-05 | 8.96E-05 |
| hsa-miR-548e-3p | 34.05684 | 13.95982 | 1.2867 | 2.22E-05 | 0.00010847 |
| hsa-miR-181c-5p | 13.52813 | 0 | 5.6551 | 2.27E-05 | 0.00011028 |
| hsa-miR-30c-1-3p | 17.59603 | 2.95304 | 2.575 | 3.33E-05 | 0.00015988 |
| hsa-miR-30d-3p | 19.86649 | 4.29533 | 2.2095 | 3.35E-05 | 0.00015996 |
| hsa-miR-200b-3p | 14.47416 | 1.342291 | 3.4307 | 3.58E-05 | 0.0001695 |
| hsa-miR-4454 | 12.96052 | 0 | 4.2714 | 4.22E-05 | 0.00019655 |
| hsa-miR-32-3p | 20.2449 | 4.832247 | 2.0668 | 4.70E-05 | 0.00021618 |
| hsa-miR-2115-3p | 12.67671 | 0 | 3.9764 | 6.29E-05 | 0.00028622 |
| hsa-miR-671-3p | 14.28495 | 1.879207 | 2.9263 | 9.10E-05 | 0.00040916 |
| hsa-miR-7-1-3p | 18.25825 | 4.29533 | 2.0877 | 0.000104 | 0.00046299 |
| hsa-miR-766-3p | 15.70399 | 2.818811 | 2.478 | 0.000112 | 0.00049842 |
| hsa-miR-2355-3p | 11.35228 | 0 | 4.4021 | 0.000119 | 0.00052583 |
| hsa-miR-1273h-3p | 13.05512 | 1.610749 | 3.0188 | 0.000157 | 0.00068653 |
| hsa-miR-193a-5p | 22.89376 | 8.053745 | 1.5072 | 0.000177 | 0.00076703 |
| hsa-miR-491-5p | 11.54148 | 1.208062 | 3.2561 | 0.000275 | 0.0011652 |
| hsa-miR-671-5p | 10.02785 | 0 | 6.2232 | 0.000308 | 0.0012841 |
| hsa-miR-3065-5p | 9.649438 | 0 | 4.5827 | 0.000365 | 0.0014968 |
| hsa-miR-338-3p | 9.649438 | 0 | 4.5827 | 0.000365 | 0.0014968 |
| hsa-miR-26b-3p | 21.28552 | 7.785286 | 1.4511 | 0.000384 | 0.001566 |
| hsa-miR-25-5p | 15.32558 | 4.563789 | 1.7476 | 0.001015 | 0.0039956 |
| hsa-miR-1255a | 8.230403 | 0 | 4.3532 | 0.001068 | 0.0041816 |
| hsa-miR-16-1-3p | 8.419607 | 0 | 3.6491 | 0.001365 | 0.0052082 |
| hsa-miR-200c-3p | 20.52871 | 8.859119 | 1.2124 | 0.00137 | 0.0052082 |
| hsa-miR-139-5p | 11.91989 | 3.221498 | 1.8876 | 0.002691 | 0.010036 |
| hsa-miR-6503-3p | 7.189777 | 0 | 3.7432 | 0.002918 | 0.010833 |
| hsa-miR-4772-5p | 6.811368 | 0 | 4.0802 | 0.003209 | 0.0118 |
| hsa-miR-4677-3p | 12.1091 | 3.892643 | 1.6373 | 0.00459 | 0.016802 |
| hsa-miR-2115-5p | 6.338356 | 0 | 3.9764 | 0.004661 | 0.016984 |
| hsa-miR-628-5p | 6.527561 | 0 | 3.6038 | 0.004941 | 0.01792 |
| hsa-miR-381-3p | 10.21705 | 2.684582 | 1.9282 | 0.005033 | 0.018087 |
| hsa-miR-296-3p | 6.338356 | 0 | 3.6641 | 0.005422 | 0.019397 |
| hsa-miR-1307-5p | 8.51421 | 2.013436 | 2.0802 | 0.008123 | 0.028538 |
| hsa-miR-33a-3p | 6.149151 | 0 | 3.1957 | 0.008281 | 0.028964 |
| hsa-miR-1250-5p | 5.959947 | 0 | 3.1506 | 0.009645 | 0.033291 |
| hsa-miR-4645-3p | 5.013923 | 0 | 3.6382 | 0.013544 | 0.045747 |
| hsa-miR-499a-5p | 12.86592 | 5.906079 | 1.1233 | 0.014552 | 0.048734 |
| Down-regulated | |||||
| hsa-let-7a-5p | 13,005.93 | 35,647.22 | −1.4546 | 0 | 0 |
| hsa-let-7b-5p | 3,975.757 | 12,791.09 | −1.6858 | 0 | 0 |
| hsa-let-7i-5p | 44,505.29 | 120,866 | −1.4414 | 0 | 0 |
| hsa-miR-25-3p | 12,852.29 | 44,411.7 | −1.7889 | 0 | 0 |
| hsa-miR-486-3p | 9,452.381 | 86,019.36 | −3.1859 | 0 | 0 |
| hsa-miR-486-5p | 9,647.451 | 87,202.59 | −3.1762 | 0 | 0 |
| hsa-miR-92a-3p | 12,505.86 | 53,627.6 | −2.1004 | 0 | 0 |
| hsa-miR-151a-3p | 788.5104 | 3,640.695 | −2.207 | 7.14E-250 | 1.65E-248 |
| hsa-miR-183-5p | 1,006.285 | 3,816.67 | −1.9233 | 5.32E-199 | 1.10E-197 |
| hsa-miR-106b-3p | 2,674.313 | 6,842.461 | −1.3553 | 3.02E-150 | 5.53E-149 |
| hsa-miR-3184-3p | 2,537.802 | 6,521.385 | −1.3616 | 3.75E-145 | 6.42E-144 |
| hsa-miR-423-5p | 2,537.802 | 6,521.385 | −1.3616 | 3.75E-145 | 6.42E-144 |
| hsa-miR-3158-5p | 217.8692 | 1,386.586 | −2.67 | 1.69E-133 | 2.72E-132 |
| hsa-miR-3158-3p | 218.7206 | 1,386.855 | −2.6647 | 4.19E-133 | 6.60E-132 |
| hsa-let-7c-5p | 535.8276 | 2,053.973 | −1.9386 | 9.96E-110 | 1.33E-108 |
| hsa-miR-30a-5p | 29.32672 | 592.7556 | −4.3371 | 1.30E-102 | 1.70E-101 |
| hsa-let-7d-5p | 837.9874 | 2,560.688 | −1.6115 | 6.17E-90 | 7.71E-89 |
| hsa-miR-3615 | 177.947 | 1,014.101 | −2.5107 | 6.64E-89 | 8.17E-88 |
| hsa-miR-144-3p | 311.7147 | 1,365.647 | −2.1313 | 7.73E-89 | 9.37E-88 |
| hsa-miR-584-5p | 67.64067 | 604.0308 | −3.1587 | 3.64E-75 | 4.15E-74 |
| hsa-miR-363-3p | 2,686.044 | 5,833.73 | −1.1189 | 2.09E-70 | 2.35E-69 |
| hsa-miR-1180-3p | 24.502 | 360.2708 | −3.8781 | 2.61E-57 | 2.81E-56 |
| hsa-miR-144-5p | 1,005.339 | 2,512.231 | −1.3213 | 2.63E-52 | 2.76E-51 |
| hsa-miR-484 | 313.0391 | 1,013.832 | −1.6954 | 3.10E-41 | 3.01E-40 |
| hsa-miR-22-3p | 344.6363 | 942.5566 | −1.4515 | 1.79E-26 | 1.55E-25 |
| hsa-miR-142-5p | 1,432.752 | 2,882.435 | −1.0085 | 1.41E-24 | 1.17E-23 |
| hsa-miR-185-3p | 43.04406 | 238.2566 | −2.4686 | 1.30E-21 | 1.02E-20 |
| hsa-miR-501-3p | 61.1131 | 285.2368 | −2.2226 | 1.98E-21 | 1.52E-20 |
| hsa-miR-7706 | 30.84036 | 137.3163 | −2.1546 | 1.53E-10 | 9.27E-10 |
| hsa-miR-2110 | 11.82529 | 74.63137 | −2.6579 | 1.33E-08 | 7.63E-08 |
| hsa-miR-5010-3p | 18.25825 | 79.32938 | −2.1193 | 1.68E-06 | 8.77E-06 |
| hsa-miR-4685-3p | 1.986649 | 30.73846 | −3.9516 | 2.56E-06 | 1.32E-05 |
| hsa-miR-4732-3p | 5.203128 | 41.61101 | −2.9995 | 3.37E-06 | 1.71E-05 |
| hsa-miR-1306-5p | 4.257105 | 34.49687 | −3.0185 | 2.14E-05 | 0.00010511 |
| hsa-miR-1299 | 0 | 19.59745 | −5.2926 | 3.33E-05 | 0.00015988 |
| hsa-miR-502-3p | 80.88499 | 197.9879 | −1.2915 | 3.70E-05 | 0.00017426 |
| hsa-miR-92b-3p | 19.01507 | 71.67833 | −1.9144 | 4.13E-05 | 0.00019337 |
| hsa-miR-320b | 61.86992 | 155.0346 | −1.3253 | 0.000148 | 0.00064948 |
| hsa-miR-629-5p | 174.7305 | 356.9151 | −1.0304 | 0.000173 | 0.00075186 |
| hsa-miR-7976 | 34.43525 | 98.65837 | −1.5186 | 0.000255 | 0.0010843 |
| hsa-miR-548ar-3p | 2.365058 | 21.87934 | −3.2096 | 0.000419 | 0.0016988 |
| hsa-miR-30a-3p | 3.405684 | 24.69815 | −2.8584 | 0.000545 | 0.0021671 |
| hsa-miR-4732-5p | 5.29773 | 28.59079 | −2.4321 | 0.001092 | 0.004256 |
| hsa-miR-6806-3p | 2.365058 | 19.8659 | −3.0703 | 0.001101 | 0.0042663 |
| hsa-miR-4326 | 23.08297 | 69.39643 | −1.588 | 0.001162 | 0.0044837 |
| hsa-miR-151a-5p | 55.05856 | 131.813 | −1.2595 | 0.001201 | 0.0046087 |
| hsa-miR-1294 | 7.095175 | 31.14115 | −2.1339 | 0.002526 | 0.0094649 |
| hsa-miR-550b-3p | 11.91989 | 41.87947 | −1.8129 | 0.003154 | 0.011652 |
| hsa-miR-550a-5p | 14.09575 | 44.42982 | −1.6563 | 0.006198 | 0.022071 |
| hsa-miR-550a-3-5p | 14.09575 | 44.2956 | −1.6519 | 0.006445 | 0.022846 |
| hsa-miR-4742-3p | 2.83807 | 16.91286 | −2.5751 | 0.008381 | 0.029184 |
| hsa-miR-18a-3p | 12.67671 | 39.59758 | −1.6432 | 0.010488 | 0.036044 |
| hsa-miR-3135a | 0 | 7.651057 | −4.7527 | 0.01163 | 0.039624 |
Table S4
| sRNA | jxzl readcount | mxzl readcount | Log2 fold change | P value | Q value |
|---|---|---|---|---|---|
| Up-regulated | |||||
| novel_581 | 43.86039 | 0 | 4.1777 | 1.60E-05 | 0.000131 |
| hsa-miR-584-3p | 106.4925 | 5.941171 | 3.669 | 3.41E-08 | 4.66E-07 |
| hsa-miR-1299 | 87.3123 | 0 | 3.4843 | 0.000621 | 0.003157 |
| hsa-miR-1180-3p | 2,554.122 | 256.4605 | 3.2663 | 9.87E-36 | 1.30E-33 |
| hsa-miR-4685-3p | 339.1172 | 20.7941 | 3.1909 | 0.000114 | 0.000726 |
| hsa-miR-3163 | 27.10556 | 1.98039 | 3.096 | 7.07E-05 | 0.000492 |
| hsa-miR-486-5p | 905,065.5 | 100,979.1 | 3.09 | 2.15E-21 | 1.24E-19 |
| hsa-miR-486-3p | 886,345.6 | 98,937.33 | 3.0893 | 2.18E-21 | 1.24E-19 |
| hsa-miR-3117-3p | 16.35697 | 0 | 3.0067 | 0.003646 | 0.014902 |
| hsa-miR-149-5p | 11.44212 | 0 | 2.9189 | 0.004822 | 0.018983 |
| hsa-miR-3135a | 29.63502 | 2.970585 | 2.8394 | 9.78E-05 | 0.000636 |
| hsa-miR-6873-3p | 7.373616 | 0 | 2.8114 | 0.006661 | 0.02412 |
| hsa-miR-4732-3p | 394.0144 | 54.46073 | 2.8016 | 2.51E-22 | 1.81E-20 |
| hsa-miR-584-5p | 4,721.776 | 707.9895 | 2.7041 | 4.73E-30 | 4.69E-28 |
| hsa-miR-1908-5p | 37.231 | 3.96078 | 2.6948 | 0.000473 | 0.002501 |
| hsa-miR-6783-3p | 27.4183 | 2.970585 | 2.6216 | 0.000951 | 0.004654 |
| hsa-miR-122-5p | 113.8024 | 5.941171 | 2.5963 | 0.008996 | 0.030964 |
| hsa-miR-3591-3p | 113.8024 | 5.941171 | 2.5963 | 0.008996 | 0.030964 |
| hsa-miR-184 | 24.20813 | 2.970585 | 2.5742 | 0.000455 | 0.002437 |
| hsa-miR-1306-5p | 304.2234 | 44.55878 | 2.5729 | 2.61E-06 | 2.44E-05 |
| hsa-miR-129-5p | 26.97631 | 1.98039 | 2.5691 | 0.006648 | 0.02412 |
| hsa-miR-6509-5p | 5.764224 | 0 | 2.5666 | 0.01454 | 0.046121 |
| hsa-miR-1224-5p | 18.85602 | 1.98039 | 2.5448 | 0.002509 | 0.01087 |
| hsa-miR-877-3p | 11.53949 | 0.990195 | 2.5421 | 0.005135 | 0.019863 |
| hsa-miR-6780a-5p | 27.21882 | 2.970585 | 2.5398 | 0.002251 | 0.009864 |
| hsa-miR-6881-3p | 5.88265 | 0 | 2.5099 | 0.017291 | 0.053146 |
| hsa-miR-6859-5p | 35.44486 | 4.950976 | 2.4945 | 0.000277 | 0.001605 |
| hsa-miR-4646-3p | 11.44282 | 0.990195 | 2.4709 | 0.007627 | 0.027235 |
| novel_133 | 6.048755 | 0 | 2.4704 | 0.019477 | 0.058949 |
| hsa-miR-1229-3p | 12.04879 | 0.990195 | 2.4626 | 0.00902 | 0.030964 |
| hsa-miR-2110 | 689.831 | 123.7744 | 2.434 | 1.54E-17 | 6.41E-16 |
| hsa-miR-4755-3p | 49.03555 | 7.921561 | 2.4166 | 3.12E-05 | 0.000235 |
| hsa-miR-5010-5p | 17.04359 | 1.98039 | 2.4105 | 0.004979 | 0.019356 |
| hsa-miR-3140-3p | 15.15428 | 1.98039 | 2.3458 | 0.004557 | 0.018069 |
| hsa-miR-129-2-3p | 5.020808 | 0 | 2.3417 | 0.027623 | 0.077403 |
| hsa-miR-6511b-5p | 4.665778 | 0 | 2.3381 | 0.027772 | 0.077546 |
| hsa-miR-3680-3p | 4.646152 | 0 | 2.3317 | 0.028257 | 0.078348 |
| hsa-miR-6743-3p | 4.487318 | 0 | 2.331 | 0.028245 | 0.078348 |
| hsa-miR-6747-3p | 4.647815 | 0 | 2.3039 | 0.030417 | 0.083176 |
| hsa-miR-4755-5p | 44.55411 | 7.921561 | 2.2917 | 7.36E-05 | 0.000503 |
| novel_692 | 9.068756 | 0.990195 | 2.2912 | 0.01216 | 0.039845 |
| hsa-miR-129-1-3p | 19.34692 | 1.98039 | 2.2864 | 0.015418 | 0.048325 |
| novel_224 | 5.001058 | 0 | 2.2278 | 0.03724 | 0.097127 |
| hsa-miR-939-5p | 22.7517 | 3.96078 | 2.2263 | 0.001039 | 0.005025 |
| hsa-miR-185-3p | 2219.59 | 450.5388 | 2.2261 | 4.97E-09 | 7.72E-08 |
| hsa-miR-92b-3p | 999.5418 | 199.0292 | 2.2184 | 1.07E-06 | 1.07E-05 |
| hsa-miR-6734-3p | 8.581901 | 0.990195 | 2.2178 | 0.016242 | 0.050709 |
| hsa-miR-4498 | 5.306282 | 0 | 2.1892 | 0.041104 | 0.10549 |
| hsa-miR-3656 | 8.299955 | 0.990195 | 2.1838 | 0.01772 | 0.054254 |
| hsa-miR-378i | 3.801219 | 0 | 2.1571 | 0.043984 | 0.11038 |
| hsa-miR-4637 | 17.64139 | 2.970585 | 2.1272 | 0.006506 | 0.023997 |
| hsa-miR-6793-3p | 4.115018 | 0 | 2.1213 | 0.048093 | 0.11918 |
| hsa-miR-370-3p | 71.66449 | 4.950976 | 2.1118 | 0.041492 | 0.1058 |
| hsa-miR-4479 | 7.398867 | 0.990195 | 2.0557 | 0.027086 | 0.076168 |
| hsa-miR-92b-5p | 51.10774 | 7.921561 | 2.0517 | 0.020225 | 0.060522 |
| hsa-miR-375 | 12.66624 | 1.98039 | 2.0366 | 0.020988 | 0.062078 |
| hsa-miR-6855-5p | 16.22223 | 2.970585 | 2.0203 | 0.010316 | 0.034663 |
| hsa-miR-92a-3p | 541,315.7 | 130,897.9 | 2.0112 | 2.58E-12 | 5.52E-11 |
| hsa-miR-18a-3p | 556.8076 | 132.6861 | 2.0052 | 6.39E-08 | 8.05E-07 |
| hsa-miR-6805-5p | 7.084954 | 0.990195 | 2.0042 | 0.03246 | 0.087554 |
| hsa-miR-6509-3p | 7.611381 | 0.990195 | 1.9674 | 0.042926 | 0.10841 |
| hsa-miR-636 | 51.83652 | 11.88234 | 1.9571 | 0.00068 | 0.003412 |
| hsa-miR-6802-3p | 10.42612 | 1.98039 | 1.9305 | 0.019609 | 0.058949 |
| hsa-miR-6741-3p | 14.18647 | 2.970585 | 1.9245 | 0.010005 | 0.033906 |
| hsa-miR-7855-5p | 14.47374 | 2.970585 | 1.9158 | 0.012365 | 0.040082 |
| hsa-miR-548at-3p | 6.400742 | 0.990195 | 1.8896 | 0.04468 | 0.11177 |
| hsa-miR-4732-5p | 238.421 | 55.45093 | 1.8838 | 0.003862 | 0.015705 |
| hsa-miR-6857-3p | 9.88382 | 1.98039 | 1.8096 | 0.036044 | 0.094961 |
| hsa-miR-1226-3p | 31.29722 | 7.921561 | 1.8015 | 0.002906 | 0.012455 |
| hsa-miR-4646-5p | 13.75534 | 2.970585 | 1.8002 | 0.025403 | 0.072204 |
| hsa-miR-6806-3p | 89.73526 | 24.75488 | 1.7855 | 8.75E-06 | 7.54E-05 |
| hsa-miR-4742-3p | 109.7316 | 29.70585 | 1.7767 | 0.000443 | 0.002392 |
| hsa-miR-484 | 11419.2 | 3,276.556 | 1.7637 | 1.08E-08 | 1.56E-07 |
| hsa-miR-1294 | 258.3273 | 74.26463 | 1.7557 | 4.82E-08 | 6.26E-07 |
| hsa-miR-3605-3p | 47.15557 | 12.87254 | 1.7311 | 0.002248 | 0.009864 |
| hsa-miR-1976 | 39.90062 | 10.89215 | 1.73 | 0.001919 | 0.008566 |
| hsa-miR-3615 | 6,182.581 | 1,862.557 | 1.7147 | 1.97E-16 | 7.10E-15 |
| hsa-miR-501-3p | 2,146.586 | 639.666 | 1.7068 | 1.31E-07 | 1.55E-06 |
| hsa-miR-6803-3p | 16.26556 | 3.96078 | 1.698 | 0.030756 | 0.083812 |
| hsa-miR-550b-3p | 395.0103 | 124.7646 | 1.6293 | 7.51E-08 | 9.16E-07 |
| hsa-miR-1255b-5p | 98.60464 | 29.70585 | 1.6257 | 0.00157 | 0.007195 |
| hsa-miR-5690 | 55.84654 | 15.84312 | 1.622 | 0.014682 | 0.046387 |
| hsa-miR-6514-5p | 14.76871 | 3.96078 | 1.6173 | 0.029293 | 0.080378 |
| hsa-miR-1273c | 23.39069 | 6.931366 | 1.6107 | 0.005521 | 0.020849 |
| hsa-miR-6511b-3p | 90.47146 | 27.72546 | 1.5922 | 0.003179 | 0.013301 |
| hsa-miR-581 | 28.43204 | 8.911756 | 1.5495 | 0.005212 | 0.020064 |
| hsa-miR-579-5p | 38.06193 | 11.88234 | 1.5484 | 0.006021 | 0.022494 |
| hsa-miR-25-3p | 404,171.9 | 134,523.9 | 1.5379 | 3.98E-05 | 0.00029 |
| hsa-miR-183-5p | 35,076.83 | 10,532.71 | 1.5191 | 0.030875 | 0.083848 |
| hsa-miR-550a-5p | 423.7288 | 147.5391 | 1.491 | 1.05E-06 | 1.07E-05 |
| hsa-miR-550a-3-5p | 422.5492 | 147.5391 | 1.4877 | 8.47E-07 | 8.83E-06 |
| hsa-miR-7706 | 904.74 | 322.8036 | 1.4756 | 7.58E-16 | 2.31E-14 |
| hsa-miR-3158-5p | 6,357.572 | 2,280.419 | 1.4654 | 1.59E-12 | 3.61E-11 |
| hsa-miR-3158-3p | 6,361.224 | 2,289.331 | 1.4607 | 1.72E-12 | 3.80E-11 |
| hsa-miR-6511a-3p | 92.2796 | 29.70585 | 1.4584 | 0.028479 | 0.078688 |
| hsa-miR-5010-3p | 559.7698 | 191.1077 | 1.4391 | 0.009518 | 0.032395 |
| hsa-miR-4746-5p | 142.2133 | 50.49995 | 1.4339 | 0.000657 | 0.003321 |
| hsa-miR-320c | 194.1517 | 70.30385 | 1.43 | 1.01E-05 | 8.51E-05 |
| hsa-miR-3940-3p | 262.6077 | 95.05873 | 1.4195 | 0.000182 | 0.001109 |
| hsa-miR-1306-3p | 30.86028 | 10.89215 | 1.4102 | 0.005302 | 0.02031 |
| hsa-miR-1292-5p | 61.2342 | 21.78429 | 1.4 | 0.006017 | 0.022494 |
| novel_401 | 70.55146 | 24.75488 | 1.3715 | 0.026575 | 0.074995 |
| hsa-miR-320b | 1,713.411 | 647.5876 | 1.3614 | 0.000229 | 0.001387 |
| hsa-let-7d-5p | 23,237.38 | 8,771.148 | 1.3572 | 0.000567 | 0.002936 |
| hsa-let-7c-5p | 15,662.8 | 5,608.465 | 1.3538 | 0.024548 | 0.070278 |
| hsa-miR-5001-3p | 41.83044 | 15.84312 | 1.3413 | 0.001923 | 0.008566 |
| hsa-miR-106b-3p | 71,400.71 | 27,991.83 | 1.3274 | 2.73E-06 | 2.46E-05 |
| hsa-miR-7976 | 909.8989 | 360.431 | 1.314 | 1.21E-06 | 1.18E-05 |
| hsa-miR-1270 | 28.18889 | 10.89215 | 1.2716 | 0.017228 | 0.053146 |
| hsa-miR-4326 | 607.1283 | 241.6076 | 1.2688 | 0.00486 | 0.018983 |
| hsa-miR-423-5p | 65,057.49 | 26,562.97 | 1.2573 | 0.000342 | 0.001936 |
| hsa-miR-3184-3p | 65,057.33 | 26,562.97 | 1.2573 | 0.000342 | 0.001936 |
| hsa-miR-144-3p | 7,839.204 | 3,262.693 | 1.2167 | 0.003187 | 0.013301 |
| hsa-let-7b-5p | 101,123.9 | 41,613.94 | 1.2137 | 0.01159 | 0.038294 |
| hsa-miR-505-5p | 53.32341 | 22.77449 | 1.1818 | 0.003936 | 0.015924 |
| novel_94 | 113.2919 | 50.49995 | 1.1332 | 0.001374 | 0.006483 |
| hsa-miR-151b | 113.9676 | 51.49015 | 1.0961 | 0.011674 | 0.038412 |
| hsa-let-7a-5p | 303,491.1 | 136,132 | 1.0813 | 0.042007 | 0.10677 |
| hsa-miR-574-5p | 383.4059 | 181.2057 | 1.0644 | 9.75E-05 | 0.000636 |
| hsa-miR-502-3p | 1,747.092 | 846.6168 | 1.0218 | 0.001298 | 0.006161 |
| Down-regulated | |||||
| hsa-miR-582-5p | 13.42211 | 3,572.624 | −6.9696 | 2.11E-18 | 9.29E-17 |
| hsa-miR-582-3p | 33.3997 | 14,490.52 | −6.6901 | 2.56E-11 | 4.84E-10 |
| hsa-miR-145-3p | 7.576002 | 715.9111 | −5.5845 | 9.02E-12 | 1.83E-10 |
| hsa-miR-424-5p | 44.14125 | 2,166.547 | −5.4852 | 3.84E-57 | 1.52E-54 |
| hsa-miR-671-5p | 1.431981 | 104.9607 | −5.3738 | 9.88E-13 | 2.30E-11 |
| hsa-miR-296-3p | 0.824955 | 66.34307 | −5.0691 | 1.01E-08 | 1.48E-07 |
| hsa-miR-2115-5p | 1.324832 | 66.34307 | −4.7385 | 2.47E-09 | 4.08E-08 |
| hsa-miR-450b-5p | 59.1493 | 1,714.028 | −4.7283 | 2.93E-42 | 5.81E-40 |
| hsa-miR-450a-5p | 153.3337 | 4112.28 | −4.6614 | 2.13E-59 | 1.69E-56 |
| hsa-miR-3065-5p | 2.368187 | 100.9999 | −4.643 | 2.65E-09 | 4.21E-08 |
| hsa-miR-338-3p | 2.368187 | 100.9999 | −4.643 | 2.65E-09 | 4.21E-08 |
| hsa-miR-542-5p | 0.442202 | 36.63722 | −4.6245 | 2.71E-06 | 2.46E-05 |
| hsa-miR-7974 | 0.399907 | 30.69605 | −4.4303 | 8.11E-06 | 7.14E-05 |
| hsa-miR-542-3p | 34.63148 | 919.8913 | −4.4264 | 3.97E-16 | 1.31E-14 |
| hsa-miR-744-3p | 0 | 17.82351 | −4.2871 | 7.44E-05 | 0.000504 |
| hsa-miR-935 | 0 | 16.83332 | −4.2012 | 0.000105 | 0.00068 |
| hsa-miR-6503-3p | 2.736968 | 75.25483 | −4.1919 | 6.83E-09 | 1.04E-07 |
| hsa-miR-145-5p | 202.251 | 8,533.502 | −4.1087 | 3.04E-05 | 0.000234 |
| hsa-miR-143-5p | 7.624172 | 279.235 | −4.0255 | 2.70E-05 | 0.00021 |
| hsa-miR-24-2-5p | 49.20377 | 972.3716 | −4.0221 | 3.09E-13 | 7.90E-12 |
| hsa-miR-4521 | 35.75842 | 647.5876 | −3.9961 | 1.49E-18 | 7.36E-17 |
| hsa-miR-652-5p | 19.04256 | 317.8526 | −3.9418 | 7.04E-29 | 6.20E-27 |
| hsa-miR-4772-5p | 1.102332 | 71.29405 | −3.9057 | 0.000268 | 0.001565 |
| hsa-miR-19b-1-5p | 0.774319 | 27.72546 | −3.9007 | 3.82E-05 | 0.00028 |
| hsa-miR-2355-3p | 3.538174 | 118.8234 | −3.898 | 4.66E-05 | 0.000336 |
| hsa-miR-181c-3p | 15.1564 | 509.9505 | −3.7733 | 0.000164 | 0.001008 |
| hsa-miR-642a-5p | 1.585413 | 32.67644 | −3.7545 | 8.06E-07 | 8.53E-06 |
| hsa-miR-642b-3p | 1.585413 | 32.67644 | −3.7545 | 8.06E-07 | 8.53E-06 |
| hsa-miR-6718-5p | 0.734962 | 23.76468 | −3.7293 | 9.02E-05 | 0.000596 |
| hsa-miR-181c-5p | 5.169999 | 141.5979 | −3.7254 | 7.36E-05 | 0.000503 |
| hsa-miR-2115-3p | 3.966267 | 132.6861 | −3.6401 | 0.000373 | 0.002042 |
| hsa-miR-551a | 1.179632 | 32.67644 | −3.5664 | 0.000263 | 0.001558 |
| hsa-miR-642a-3p | 0.399907 | 16.83332 | −3.5654 | 0.00062 | 0.003157 |
| hsa-miR-642b-5p | 0.399907 | 16.83332 | −3.5654 | 0.00062 | 0.003157 |
| hsa-miR-26a-1-3p | 0.623711 | 17.82351 | −3.5259 | 0.000359 | 0.001977 |
| hsa-miR-1537-5p | 0.388628 | 15.84312 | −3.5096 | 0.000766 | 0.003798 |
| hsa-miR-34c-5p | 17.37958 | 421.8231 | −3.5 | 0.000349 | 0.00196 |
| hsa-miR-3065-3p | 26.58372 | 603.0288 | −3.4811 | 0.000284 | 0.001632 |
| hsa-miR-429 | 0.226741 | 14.85293 | −3.4595 | 0.001384 | 0.006494 |
| hsa-miR-769-5p | 189.6205 | 2130.9 | −3.4522 | 4.46E-52 | 1.18E-49 |
| hsa-miR-874-3p | 14.487 | 206.9508 | −3.4015 | 1.91E-06 | 1.81E-05 |
| hsa-miR-338-5p | 26.39928 | 600.0582 | −3.35 | 0.000794 | 0.003909 |
| hsa-miR-191-3p | 2.393459 | 47.52937 | −3.3379 | 0.00041 | 0.002225 |
| hsa-miR-4454 | 9.186323 | 135.6567 | −3.3321 | 1.75E-05 | 0.00014 |
| hsa-miR-548w | 1.726045 | 33.66663 | −3.3084 | 0.000546 | 0.00287 |
| hsa-miR-2355-5p | 0.161887 | 11.88234 | −3.2833 | 0.002453 | 0.010688 |
| hsa-miR-28-5p | 41.97051 | 421.8231 | −3.2771 | 8.42E-31 | 9.54E-29 |
| hsa-miR-5091 | 0.21546 | 12.87254 | −3.2707 | 0.00256 | 0.011032 |
| hsa-miR-181b-3p | 2.140029 | 49.50976 | −3.2254 | 0.001504 | 0.006974 |
| hsa-miR-9-3p | 1.33905 | 21.78429 | −3.1872 | 0.000354 | 0.00196 |
| hsa-miR-199a-3p | 706.5011 | 8,051.277 | −3.179 | 1.17E-06 | 1.16E-05 |
| hsa-miR-4286 | 23.80597 | 309.9311 | −3.1773 | 4.78E-05 | 0.000342 |
| hsa-miR-33a-3p | 6.041826 | 64.36268 | −3.1238 | 1.98E-07 | 2.28E-06 |
| hsa-miR-24-3p | 2,255.433 | 21006 | −3.1187 | 3.21E-16 | 1.11E-14 |
| hsa-miR-873-5p | 0.442202 | 12.87254 | −3.1166 | 0.003411 | 0.014087 |
| hsa-miR-618 | 80.56582 | 1,795.224 | −3.1115 | 0.002932 | 0.0125 |
| hsa-miR-3074-5p | 2,252.299 | 20,805.98 | −3.1071 | 4.35E-16 | 1.38E-14 |
| hsa-miR-4791 | 0 | 7.921561 | −3.0996 | 0.003647 | NA |
| hsa-miR-340-5p | 1,420.416 | 12,837.88 | −3.0855 | 2.57E-17 | 1.02E-15 |
| hsa-miR-200b-3p | 14.76434 | 151.4999 | −3.085 | 1.92E-07 | 2.24E-06 |
| hsa-miR-27a-3p | 1,623.284 | 15,193.55 | −3.0228 | 2.05E-08 | 2.85E-07 |
| hsa-miR-30b-5p | 1,635.638 | 14,427.14 | −3.0227 | 4.88E-13 | 1.17E-11 |
| hsa-miR-146b-5p | 2,804.876 | 25,328.2 | −3.0225 | 1.38E-10 | 2.49E-09 |
| hsa-miR-140-5p | 241.5527 | 2,217.047 | −2.993 | 3.60E-08 | 4.84E-07 |
| hsa-miR-223-5p | 452.9507 | 4,237.045 | −2.9857 | 3.01E-07 | 3.36E-06 |
| hsa-miR-628-5p | 7.574612 | 68.32346 | −2.97 | 3.84E-08 | 5.08E-07 |
| hsa-miR-330-3p | 33.61209 | 288.1468 | −2.9638 | 1.05E-11 | 2.08E-10 |
| hsa-miR-16-1-3p | 10.5328 | 88.12737 | −2.94 | 7.77E-12 | 1.62E-10 |
| hsa-miR-23a-5p | 0.346333 | 9.901951 | −2.9281 | 0.006042 | 0.022494 |
| hsa-miR-651-5p | 157.9455 | 1,220.911 | −2.921 | 1.10E-40 | 1.74E-38 |
| hsa-miR-23a-3p | 4,204.238 | 34,490.48 | −2.886 | 1.58E-09 | 2.72E-08 |
| hsa-miR-7850-5p | 0.161887 | 8.911756 | −2.8796 | 0.007948 | NA |
| hsa-miR-3690 | 3.245414 | 50.49995 | −2.8703 | 0.004361 | 0.017376 |
| hsa-miR-556-3p | 0.399907 | 9.901951 | −2.8366 | 0.008139 | 0.028687 |
| hsa-miR-148a-5p | 165.23 | 1,273.391 | −2.7884 | 1.98E-08 | 2.80E-07 |
| hsa-miR-491-5p | 11.303 | 120.8038 | −2.7851 | 0.001393 | 0.006496 |
| hsa-miR-222-3p | 1,908.206 | 14,124.14 | −2.73 | 5.10E-08 | 6.52E-07 |
| hsa-miR-371a-3p | 0 | 5.941171 | −2.7068 | 0.00926 | NA |
| hsa-miR-371b-5p | 0 | 5.941171 | −2.7068 | 0.00926 | NA |
| hsa-miR-3917 | 0 | 5.941171 | −2.7068 | 0.00926 | NA |
| hsa-miR-7704 | 4.324936 | 49.50976 | −2.7047 | 0.003991 | 0.016066 |
| hsa-miR-365b-5p | 1.244486 | 14.85293 | −2.6965 | 0.005431 | 0.020605 |
| hsa-miR-21-5p | 45,063.79 | 296,309.9 | −2.6724 | 5.87E-22 | 3.88E-20 |
| hsa-miR-335-3p | 25.81005 | 182.1959 | −2.6463 | 2.54E-07 | 2.88E-06 |
| hsa-miR-3667-3p | 3.85645 | 47.52937 | −2.6408 | 0.008541 | 0.029969 |
| hsa-miR-4645-3p | 7.262121 | 52.48034 | −2.6395 | 8.32E-06 | 7.25E-05 |
| hsa-miR-181a-3p | 243.0981 | 2,664.615 | −2.6036 | 0.007672 | 0.027235 |
| hsa-miR-4477b | 0.223804 | 7.921561 | −2.5641 | 0.017468 | NA |
| hsa-miR-1537-3p | 0.732024 | 9.901951 | −2.5587 | 0.012384 | 0.040082 |
| hsa-miR-708-5p | 0.447607 | 16.83332 | −2.5474 | 0.016906 | 0.052369 |
| hsa-miR-4676-5p | 1.954418 | 16.83332 | −2.5298 | 0.003158 | 0.013301 |
| hsa-miR-4773 | 0.677286 | 12.87254 | −2.5287 | 0.019625 | 0.058949 |
| hsa-miR-142-3p | 2844.32 | 17,667.06 | −2.5263 | 7.07E-09 | 1.06E-07 |
| novel_498 | 1.213583 | 11.88234 | −2.5259 | 0.007475 | 0.026824 |
| hsa-miR-548ag | 0.592808 | 10.89215 | −2.4986 | 0.020813 | 0.062049 |
| hsa-miR-26a-2-3p | 5.624783 | 45.54898 | −2.4823 | 0.003472 | 0.014266 |
| hsa-miR-34c-3p | 0 | 4.950976 | −2.4713 | 0.015201 | NA |
| hsa-miR-3607-3p | 0 | 4.950976 | −2.4713 | 0.015201 | NA |
| hsa-miR-511-3p | 0 | 4.950976 | −2.4713 | 0.015201 | NA |
| hsa-miR-873-3p | 0 | 4.950976 | −2.4713 | 0.015201 | NA |
| hsa-miR-7-1-3p | 33.35981 | 191.1077 | −2.4617 | 1.09E-14 | 3.10E-13 |
| hsa-miR-424-3p | 348.9398 | 1,957.616 | −2.4287 | 3.22E-13 | 7.99E-12 |
| hsa-miR-1273h-5p | 3.191369 | 33.66663 | −2.427 | 0.016466 | 0.051207 |
| hsa-miR-6839-5p | 0.734962 | 8.911756 | −2.422 | 0.018759 | 0.057214 |
| hsa-miR-1248 | 0.385691 | 6.931366 | −2.411 | 0.025824 | NA |
| hsa-miR-361-5p | 175.5968 | 968.4108 | −2.4095 | 4.78E-14 | 1.31E-12 |
| hsa-miR-27b-3p | 4,443.556 | 24,239.98 | −2.4087 | 1.12E-18 | 5.91E-17 |
| hsa-miR-4677-5p | 0.21546 | 6.931366 | −2.4059 | 0.025053 | NA |
| hsa-miR-200a-5p | 2.172235 | 18.81371 | −2.3983 | 0.010665 | 0.035595 |
| hsa-miR-450a-2-3p | 4.107031 | 25.74507 | −2.3963 | 0.000133 | 0.000838 |
| hsa-miR-221-3p | 3,125.573 | 17,615.57 | −2.3877 | 7.74E-08 | 9.30E-07 |
| hsa-miR-197-3p | 895.456 | 5,296.554 | −2.3782 | 3.33E-05 | 0.000249 |
| hsa-miR-1291 | 2.002116 | 15.84312 | −2.3692 | 0.008697 | 0.03025 |
| hsa-miR-28-3p | 1,214.146 | 6,413.494 | −2.3642 | 1.63E-18 | 7.59E-17 |
| hsa-miR-21-3p | 57.77555 | 312.9017 | −2.353 | 2.37E-09 | 3.99E-08 |
| hsa-miR-1273h-3p | 22.35517 | 136.6469 | −2.3474 | 0.000466 | 0.002478 |
| hsa-miR-590-5p | 1.162945 | 9.901951 | −2.3286 | 0.014167 | 0.045119 |
| hsa-miR-32-3p | 37.8722 | 211.9018 | −2.314 | 2.35E-05 | 0.000187 |
| hsa-miR-9-5p | 86.99068 | 479.2544 | −2.3072 | 1.59E-05 | 0.000131 |
| hsa-miR-1307-5p | 16.08154 | 89.11756 | −2.2999 | 1.33E-05 | 0.000111 |
| hsa-miR-27b-5p | 5.909069 | 34.65683 | −2.2962 | 0.000244 | 0.001463 |
| hsa-miR-155-5p | 1,343.81 | 8,091.875 | −2.2667 | 0.002114 | 0.009364 |
| hsa-miR-365a-3p | 41.40705 | 239.6272 | −2.2575 | 0.001017 | 0.00495 |
| hsa-miR-30d-3p | 40.67332 | 207.941 | −2.2269 | 9.57E-06 | 8.16E-05 |
| hsa-miR-29a-3p | 1,581.287 | 7,876.012 | −2.2085 | 1.81E-06 | 1.75E-05 |
| hsa-miR-190b | 9.734646 | 49.50976 | −2.2029 | 5.34E-05 | 0.000378 |
| hsa-miR-548aw | 0 | 3.96078 | −2.1954 | 0.025861 | NA |
| hsa-miR-6503-5p | 0 | 3.96078 | −2.1954 | 0.025861 | NA |
| novel_377 | 0 | 3.96078 | −2.1954 | 0.025861 | NA |
| novel_580 | 0 | 3.96078 | −2.1954 | 0.025861 | NA |
| hsa-miR-885-5p | 0.399907 | 5.941171 | −2.19 | 0.043508 | NA |
| hsa-miR-222-5p | 0.223804 | 5.941171 | −2.1897 | 0.03961 | NA |
| hsa-miR-34b-3p | 0.223804 | 5.941171 | −2.1897 | 0.03961 | NA |
| hsa-miR-1250-5p | 7.483318 | 62.38229 | −2.1719 | 0.033298 | 0.089509 |
| hsa-miR-200a-3p | 0.670107 | 9.901951 | −2.1461 | 0.048002 | 0.11918 |
| hsa-miR-3609 | 0.161887 | 4.950976 | −2.1186 | 0.045782 | NA |
| hsa-miR-3154 | 1.100917 | 9.901951 | −2.1171 | 0.04135 | 0.10578 |
| hsa-miR-6854-5p | 0.732024 | 6.931366 | −2.1126 | 0.0433 | NA |
| hsa-miR-548au-5p | 0.765976 | 6.931366 | −2.0772 | 0.047845 | NA |
| hsa-miR-616-5p | 2.522921 | 14.85293 | −2.0614 | 0.021859 | 0.063328 |
| hsa-miR-556-5p | 1.672469 | 11.88234 | −2.052 | 0.040919 | 0.10535 |
| hsa-miR-30c-1-3p | 43.47084 | 184.1763 | −2.0458 | 2.98E-13 | 7.89E-12 |
| hsa-miR-514a-3p | 6.992634 | 35.64702 | −2.039 | 0.007693 | 0.027235 |
| hsa-miR-152-3p | 134.0388 | 595.1073 | −2.0272 | 8.33E-05 | 0.000555 |
| hsa-miR-548f-3p | 13.37504 | 60.4019 | −2.0206 | 0.000267 | 0.001565 |
| hsa-miR-92a-1-5p | 5.582602 | 28.71566 | −2.0166 | 0.008664 | 0.03025 |
| hsa-miR-671-3p | 33.07217 | 149.5195 | −2.0092 | 0.000555 | 0.002897 |
| hsa-miR-20a-5p | 12,399.55 | 50,294.98 | −1.9812 | 3.89E-11 | 7.18E-10 |
| hsa-miR-29c-3p | 78.70773 | 331.7154 | −1.9672 | 6.08E-05 | 0.000427 |
| hsa-miR-374b-3p | 58.09844 | 233.686 | −1.9213 | 2.69E-05 | 0.00021 |
| hsa-miR-548aq-3p | 4.145114 | 19.8039 | −1.9024 | 0.021229 | 0.062119 |
| hsa-miR-548g-5p | 4.145114 | 19.8039 | −1.9024 | 0.021229 | 0.062119 |
| hsa-miR-340-3p | 168.3928 | 675.3131 | −1.8992 | 0.00011 | 0.000705 |
| hsa-miR-193a-5p | 58.48551 | 239.6272 | −1.8933 | 0.000686 | 0.003421 |
| hsa-miR-26a-5p | 84,097.22 | 316,405 | −1.8906 | 8.43E-17 | 3.19E-15 |
| hsa-miR-216b-5p | 0 | 2.970585 | −1.8628 | 0.046126 | NA |
| hsa-miR-3186-3p | 0 | 2.970585 | −1.8628 | 0.046126 | NA |
| hsa-miR-548u | 6.304767 | 27.72546 | −1.8509 | 0.013435 | 0.043309 |
| hsa-miR-148a-3p | 170,162.7 | 736,105.1 | −1.8458 | 0.012368 | 0.040082 |
| hsa-miR-188-5p | 3.107743 | 14.85293 | −1.8315 | 0.038203 | 0.099003 |
| hsa-miR-345-5p | 330.4409 | 1,256.558 | −1.8294 | 0.000155 | 0.000962 |
| hsa-miR-30c-5p | 10,728.19 | 38,116.57 | −1.78 | 4.34E-07 | 4.78E-06 |
| hsa-miR-103a-2-5p | 5.75189 | 22.77449 | −1.7702 | 0.009288 | 0.031748 |
| hsa-miR-126-5p | 373.3419 | 1,270.42 | −1.7558 | 8.30E-23 | 6.58E-21 |
| hsa-miR-146a-5p | 2,754.016 | 9,687.079 | −1.726 | 0.000256 | 0.001525 |
| hsa-miR-19b-3p | 742.0626 | 2,678.478 | −1.7138 | 0.003015 | 0.012783 |
| hsa-miR-20a-3p | 12.45507 | 42.57839 | −1.6239 | 0.011311 | 0.037531 |
| hsa-miR-2277-5p | 26.6079 | 85.15678 | −1.5819 | 0.001896 | 0.008544 |
| hsa-miR-106b-5p | 2,927.107 | 8,687.972 | −1.537 | 7.60E-07 | 8.26E-06 |
| hsa-miR-3912-3p | 9.738392 | 30.69605 | −1.5366 | 0.006537 | 0.023999 |
| hsa-miR-502-5p | 5.100938 | 16.83332 | −1.5256 | 0.034766 | 0.092372 |
| hsa-miR-4677-3p | 41.77458 | 126.745 | −1.525 | 0.001623 | 0.007395 |
| hsa-miR-548k | 114.4978 | 331.7154 | −1.5044 | 1.90E-06 | 1.81E-05 |
| hsa-miR-362-5p | 290.0476 | 839.6855 | −1.5042 | 9.85E-07 | 1.01E-05 |
| hsa-miR-128-1-5p | 12.37597 | 40.598 | −1.4763 | 0.04988 | 0.12322 |
| hsa-miR-425-3p | 840.5069 | 2,319.037 | −1.4477 | 3.86E-10 | 6.80E-09 |
| hsa-miR-301a-3p | 5.658604 | 16.83332 | −1.4163 | 0.02372 | 0.068153 |
| hsa-miR-660-5p | 608.0902 | 1,641.744 | −1.4123 | 7.45E-08 | 9.16E-07 |
| hsa-miR-4676-3p | 11.00521 | 31.68624 | −1.4079 | 0.02101 | 0.062078 |
| hsa-miR-766-3p | 57.79367 | 164.3724 | −1.4018 | 0.01359 | 0.043631 |
| hsa-miR-505-3p | 234.728 | 635.7053 | −1.3995 | 8.10E-05 | 0.000544 |
| hsa-miR-548e-3p | 134.369 | 356.4702 | −1.3947 | 1.70E-11 | 3.29E-10 |
| hsa-miR-361-3p | 5,529.697 | 14,499.43 | −1.3654 | 2.68E-06 | 2.46E-05 |
| hsa-miR-146b-3p | 208.7207 | 581.2445 | −1.3395 | 0.03542 | 0.093626 |
| hsa-miR-30e-3p | 6,326.623 | 16,068.89 | −1.3155 | 3.51E-05 | 0.00026 |
| hsa-miR-26b-3p | 87.00414 | 222.7939 | −1.2973 | 0.003402 | 0.014087 |
| hsa-miR-576-3p | 301.8402 | 712.9405 | −1.2344 | 2.77E-15 | 8.15E-14 |
| hsa-miR-200c-3p | 85.86659 | 214.8723 | −1.2132 | 0.044862 | 0.11187 |
| hsa-miR-3128 | 10.75696 | 25.74507 | −1.1715 | 0.034829 | 0.092372 |
| hsa-miR-191-5p | 49,040.71 | 110,790 | −1.1596 | 4.72E-06 | 4.20E-05 |
| hsa-miR-425-5p | 9,743.394 | 22,378.41 | −1.1571 | 0.004096 | 0.016406 |
| hsa-miR-126-3p | 28,628.66 | 64,706.28 | −1.157 | 3.10E-05 | 0.000235 |
| hsa-let-7b-3p | 30.72418 | 72.28424 | −1.1386 | 0.043703 | 0.11002 |
| hsa-miR-26b-5p | 44,744.76 | 99,966.14 | −1.1279 | 0.001525 | 0.007033 |
| hsa-miR-30e-5p | 7,858.349 | 17,603.69 | −1.1157 | 0.010071 | 0.033984 |
| hsa-miR-15a-5p | 1,351.35 | 3,040.889 | −1.1155 | 0.015379 | 0.048325 |
| hsa-miR-374b-5p | 6,150.1 | 13,442.89 | −1.1004 | 0.001107 | 0.005319 |
| hsa-miR-374c-3p | 6,146.04 | 13432 | −1.1001 | 0.001119 | 0.005346 |
| hsa-miR-590-3p | 76.42887 | 165.3626 | −1.0726 | 0.00643 | 0.023827 |
| hsa-miR-548ay-3p | 15.04833 | 32.67644 | −1.0554 | 0.031173 | 0.08437 |
| hsa-miR-6842-3p | 285.8855 | 601.0484 | −1.0175 | 0.034064 | 0.09126 |
| hsa-miR-101-3p | 56,534.22 | 114,797.3 | −1.0045 | 0.000352 | 0.00196 |
| hsa-miR-25-5p | 78.24413 | 160.4116 | −1.0028 | 0.00687 | 0.024762 |
Table S5
| sRNA | sxzl readcount | Control readcount | Log2 fold change | P value | Q value |
|---|---|---|---|---|---|
| Up-regulated | |||||
| hsa-miR-1255a | 73.93453 | 3.670145 | 2.4982 | 0.000734 | 0.98318 |
| hsa-miR-99b-3p | 34.05871 | 2.446764 | 1.8701 | 0.015512 | 0.98318 |
| hsa-miR-342-3p | 1,564.196 | 274.0375 | 1.7191 | 0.015543 | 0.98318 |
| hsa-miR-34a-5p | 144.9126 | 12.23382 | 1.8276 | 0.017862 | 0.98318 |
| hsa-miR-181a-3p | 1,231.747 | 58.72232 | 1.7736 | 0.023952 | 0.98318 |
| hsa-miR-181b-5p | 1,733.788 | 261.8037 | 1.6401 | 0.028975 | 0.98318 |
| hsa-miR-125a-5p | 565.7181 | 50.15865 | 1.6352 | 0.036442 | 0.98318 |
| hsa-miR-8485 | 12.10476 | 0 | 1.6302 | 0.038292 | 0.98318 |
| hsa-miR-99b-5p | 552.4379 | 34.25469 | 1.575 | 0.045443 | 0.98318 |
| Down-regulated | |||||
| hsa-miR-30a-5p | 554.4653 | 5,402.454 | −2.1709 | 0.004414 | 0.98318 |
| hsa-miR-30a-3p | 41.46146 | 225.1022 | −1.5811 | 0.038759 | 0.98318 |
Table S6
| sRNA | lxzl readcount | Control readcount | Log2 fold change | P value | Q value |
|---|---|---|---|---|---|
| Up-regulated | |||||
| hsa-miR-125a-5p | 164.4959262 | 42.54988 | 1.7568 | 7.52E-09 | 3.92E-06 |
| hsa-miR-1255a | 47.68217947 | 3.113406 | 2.566 | 2.19E-08 | 5.71E-06 |
| hsa-let-7e-5p | 196.18214 | 71.60834 | 1.309 | 1.89E-05 | 0.00222 |
| hsa-miR-99b-5p | 228.78698 | 29.05846 | 1.992 | 2.13E-05 | 0.00222 |
| hsa-miR-99b-3p | 23.73255619 | 2.075604 | 1.8339 | 0.000253 | 0.020466 |
| hsa-miR-34a-5p | 226.0002671 | 10.37802 | 1.8112 | 0.000441 | 0.028716 |
| hsa-miR-127-3p | 140.8358884 | 21.79384 | 1.6454 | 0.000785 | 0.036749 |
| hsa-miR-1270 | 26.93738349 | 6.226812 | 1.4834 | 0.001185 | 0.044086 |
| hsa-miR-139-5p | 175.0936703 | 24.90725 | 1.5811 | 0.001626 | 0.056467 |
| hsa-miR-493-3p | 20.24950193 | 1.037802 | 1.5177 | 0.00335 | 0.096951 |
| hsa-miR-493-5p | 36.66853096 | 5.18901 | 1.3349 | 0.009469 | 0.22425 |
| hsa-miR-143-5p | 7.137243546 | 0 | 1.2882 | 0.011921 | NA |
| hsa-miR-1226-3p | 28.83038399 | 9.340218 | 1.1487 | 0.012641 | 0.27767 |
| hsa-miR-335-3p | 18.89945023 | 5.18901 | 1.1775 | 0.015629 | 0.32382 |
| hsa-miR-1303 | 108.0305569 | 36.32307 | 1.1114 | 0.01616 | 0.32382 |
| hsa-miR-7854-3p | 6.459986627 | 0 | 1.2208 | 0.016689 | NA |
| hsa-miR-379-5p | 15.65690359 | 1.037802 | 1.2222 | 0.017663 | 0.32865 |
| hsa-miR-3656 | 8.119232076 | 1.037802 | 1.1662 | 0.0242 | NA |
| hsa-miR-6786-3p | 6.151685313 | 0 | 1.14 | 0.024322 | NA |
| hsa-miR-381-3p | 97.77120798 | 20.75604 | 1.0721 | 0.037705 | 0.49031 |
| hsa-miR-182-3p | 10.42158481 | 2.075604 | 1.0584 | 0.040467 | 0.49031 |
| hsa-miR-4433b-5p | 174.713051 | 18.68044 | 1.0067 | 0.048513 | 0.52705 |
| hsa-miR-4433a-3p | 175.0375348 | 18.68044 | 1.0055 | 0.048736 | 0.52705 |
| Down-regulated | |||||
| hsa-miR-450a-5p | 135.4842658 | 343.5125 | −1.2519 | 1.49E-06 | 0.000259 |
| hsa-miR-3690 | 2.625944012 | 28.02066 | −1.7583 | 0.000675 | 0.036749 |
| hsa-miR-3135a | 22.32185019 | 59.15472 | −1.1441 | 0.004872 | 0.13359 |
| hsa-miR-642a-5p | 1.308257088 | 11.41582 | −1.3814 | 0.00686 | NA |
| hsa-miR-642b-3p | 1.308257088 | 11.41582 | −1.3814 | 0.00686 | NA |
| hsa-miR-4435 | 5.142299703 | 18.68044 | −1.0286 | 0.046412 | 0.52705 |
Table S7
| sRNA | sxzl readcount | lxzl readcount | Log2 fold change | P value | Q value |
|---|---|---|---|---|---|
| Up-regulated | |||||
| hsa-miR-181a-5p | 39,056.69 | 15,625.02 | 1.2746 | 1.92E-06 | 0.000148 |
| hsa-miR-181a-3p | 1,215.375 | 77.99342 | 2.9279 | 3.03E-06 | 0.000148 |
| hsa-miR-181b-5p | 1,693.275 | 224.783 | 2.4416 | 4.07E-06 | 0.000148 |
| hsa-miR-342-3p | 1,523.522 | 257.5529 | 2.2109 | 8.51E-06 | 0.000232 |
| hsa-miR-146b-3p | 401.1346 | 140.4814 | 1.3875 | 0.000492 | NA |
| hsa-miR-15b-3p | 2,750.452 | 1,008.29 | 1.2939 | 0.003621 | 0.078929 |
| hsa-miR-16-2-3p | 1,296.128 | 381.1239 | 1.4855 | 0.004811 | 0.084379 |
| hsa-miR-1260b | 76.25221 | 24.49384 | 1.3869 | 0.007796 | NA |
| hsa-miR-1-3p | 90.20319 | 18.62308 | 1.6595 | 0.009336 | NA |
| hsa-miR-1268a | 11.95946 | 2.560318 | 1.63 | 0.009398 | NA |
| hsa-miR-150-5p | 2,148.384 | 716.1593 | 1.3371 | 0.010691 | 0.12348 |
| hsa-miR-197-3p | 2,577.615 | 680.3299 | 1.5043 | 0.011328 | 0.12348 |
| hsa-let-7f-1-3p | 19.77389 | 5.190162 | 1.4708 | 0.015771 | NA |
| hsa-miR-200a-3p | 4.417496 | 0 | 1.7056 | 0.016799 | NA |
| hsa-miR-1307-5p | 44.33103 | 12.17739 | 1.4361 | 0.017191 | NA |
| hsa-miR-504-5p | 31.89215 | 4.118537 | 1.6451 | 0.021032 | NA |
| hsa-miR-4707-3p | 19.32852 | 3.991825 | 1.5099 | 0.025443 | NA |
| hsa-miR-92a-1-5p | 15.18709 | 3.689128 | 1.4387 | 0.026903 | NA |
| hsa-miR-27b-5p | 16.97949 | 4.421235 | 1.3786 | 0.033286 | NA |
| hsa-miR-3154 | 4.762506 | 0.373006 | 1.5324 | 0.033713 | NA |
| hsa-miR-6770-5p | 2.865023 | 0 | 1.4812 | 0.035719 | NA |
| hsa-miR-1275 | 5.691972 | 0 | 1.4454 | 0.037007 | NA |
| hsa-miR-23a-5p | 3.877873 | 0 | 1.4563 | 0.037146 | NA |
| hsa-miR-125a-5p | 543.5221 | 187.4418 | 1.2164 | 0.037688 | NA |
| hsa-miR-499a-5p | 126.6113 | 41.07934 | 1.2463 | 0.040416 | NA |
| hsa-miR-499b-3p | 125.6438 | 41.07934 | 1.2387 | 0.041507 | NA |
| hsa-miR-125b-2-3p | 8.459143 | 1.865813 | 1.3897 | 0.042802 | NA |
| hsa-miR-125a-3p | 12.62621 | 1.487911 | 1.4403 | 0.045977 | NA |
| hsa-miR-32-3p | 101.2775 | 32.47671 | 1.2287 | 0.048942 | NA |
| Down-regulated | |||||
| hsa-miR-30a-5p | 529.2648 | 1,956.122 | −1.4991 | 0.009967 | 0.12348 |
| hsa-miR-6753-3p | 0.869689 | 5.90425 | −1.7068 | 0.013047 | NA |
| hsa-miR-151a-3p | 10,040.77 | 22,962.51 | −1.0725 | 0.013772 | 0.13647 |
| hsa-miR-3133 | 15.02586 | 34.93526 | −1.0734 | 0.020626 | NA |
| hsa-miR-1288-3p | 2.661609 | 9.564785 | −1.3787 | 0.026051 | NA |
| hsa-miR-4753-5p | 0 | 2.201217 | −1.5165 | 0.031505 | NA |
| hsa-miR-30c-2-3p | 0.269812 | 3.661317 | −1.5266 | 0.034302 | NA |
| hsa-miR-449c-5p | 5.607974 | 13.67431 | −1.0701 | 0.042598 | NA |
| hsa-miR-6815-5p | 6.328357 | 14.35981 | −1.0136 | 0.046006 | NA |
| hsa-miR-1303 | 53.95024 | 123.3373 | −1.0132 | 0.048477 | NA |
Acknowledgments
Funding: This research was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.01.18). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all patients. The study was approved by the Tumor Hospital in Yunnan Province (2015FB072).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kusenda B, Mraz M, Mayer J, et al. MicroRNA biogenesis, functionality and cancer relevance. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 2006;150:205-15. [Crossref] [PubMed]
- Griffiths-Jones S, Grocock RJ, van Dongen S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 2006;34:D140-4. [Crossref] [PubMed]
- Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92-105. [Crossref] [PubMed]
- Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol 2007;8:23-36. [Crossref] [PubMed]
- Hammell CM. The microRNA-argonaute complex: a platform for mRNA modulation. RNA Biol 2008;5:123-7. [Crossref] [PubMed]
- Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 2008;30:460-71. [Crossref] [PubMed]
- Tsuchiya S, Okuno Y, Tsujimoto G. MicroRNA: biogenetic and functional mechanisms and involvements in cell differentiation and cancer. J Pharmacol Sci 2006;101:267-70. [Crossref] [PubMed]
- Tang JT, Fang JY. MicroRNA regulatory network in human colorectal cancer. Mini Rev Med Chem 2009;9:921-6. [Crossref] [PubMed]
- Sassen S, Miska EA, Caldas C. MicroRNA: implications for cancer. Virchows Arch 2008;452:1-10. [Crossref] [PubMed]
- Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov 2013;12:847-65. [Crossref] [PubMed]
- Larsen MT, Häger M, Glenthøj A, et al. miRNA-130a regulates C/EBP-ε expression during granulopoiesis. Blood 2014;123:1079-89. [Crossref] [PubMed]
- Lee YG, Kim I, Oh S, et al. Small RNA sequencing profiles of mir-181 and mir-221, the most relevant microRNAs in acute myeloid leukemia. Korean J Intern Med 2019;34:178-83. [Crossref] [PubMed]
- Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med 2016;374:2209-21. [Crossref] [PubMed]
- Schee K, Lorenz S, Worren MM, et al. Deep Sequencing the MicroRNA Transcriptome in Colorectal Cancer. PLoS One 2013;8:e66165. [Crossref] [PubMed]
- Vaz C, Ahmad HM, Sharma P, et al. Analysis of microRNA transcriptome by deep sequencing of small RNA libraries of peripheral blood. BMC Genomics 2010;11:288. [Crossref] [PubMed]
- Wen M, Shen Y, Shi S, et al. miREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinformatics. 2012;13:140. [Crossref] [PubMed]
- Friedländer MR, Mackowiak SD, Li N, et al. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 2012;40:37-52. [Crossref] [PubMed]
- Garzon R, Volinia S, Liu CG, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 2008;111:3183-9. [Crossref] [PubMed]
- Dixon-McIver A, East P, Mein CA, et al. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One 2008;3:e2141. [Crossref] [PubMed]
- Luo HC, Zhang ZZ, Zhang X, et al. MicroRNA expression signature in gastric cancer. Chin J Cancer Res 2009;21:74-80. [Crossref]
- Navarro A, Gaya A, Martinez A, et al. MicroRNA expression profiling in classic Hodgkin lymphoma. Blood 2008;111:2825-32. [Crossref] [PubMed]
- Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 2002;99:15524-9. [Crossref] [PubMed]
- Liang Y, Ridzon D, Wong L, et al. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 2007;8:166. [Crossref] [PubMed]