
The biological functions of LGR5 in promoting non-small cell lung cancer progression
Introduction
Lung cancer is the leading cause of cancer mortality in men and the second leading cause of cancer mortality in women, and accounted for 18.4% of total global cancer mortalities in 2018 (1). With increasing pollution (2) and overabundance of fine ambient particles (PM2.5) (3), lung cancer poses significant public health problems in China, especially non-small cell lung cancer (NSCLC), which accounts for more than 80% of all lung cancers (4). Due to the poor prognosis and lack of an effective early diagnostic method for NSCLC, new targets for diagnosis and treatment are urgently needed (5).
Evidence has shown that the presence of cancer stem cells (CSCs) is one of the reasons for the poor therapeutic effect (6,7). A recent study indicated that NSCLC also harbored CSC populations, which can lead to tumor recurrence, therapy resistance, and distant metastasis (8-11).
Leucine-rich repeat-containing G-protein-coupled receptor 5 (LGR5) is a member of the G-protein-coupled receptor superfamily, which has been proven to be a stem cell marker in colorectal cancers (CRC) (12) and a potentiator of the Wnt signaling pathway (13,14). It was reported that LGR5 had a strong correlation with tumor growth, invasion, and poor prognosis in CRC. Also, liver metastases derived from the primary CRCs were dependent on Lgr5+ CSCs (15-17). LGR5+ cells in intestinal adenomas were also found to be highly proliferative and correlated with tumor expansion and invasion (18). A previous study found that LGR5 was also expressed in NSCLC and was related to cancer progression (19). However, the mechanism for LGR5 in the modulation of the biological function of NSCLC cells remains unknown. Therefore, the goal of this study was to analyze LGR5 expression in human NSCLC tissues and evaluate the relationship between LGR5 expression and clinical parameters. We also investigated the role of LGR5 in the proliferation, migration, and invasion of NSCLC in vitro.
Methods
Clinical samples
A total of 22 NSCLC tissues and matched adjacent normal tissues were collected from January 2015 to March 2017 at the Fifth People’s Hospital of Suzhou (Suzhou, China). None of the patients had received preoperative chemotherapy or radiotherapy. The median age was 64 years (range, 42–77 years), and the sample included 17 males and 5 females. Disease stage was classified according to the criteria recommended by the International Association for the Study of Lung Cancer (IASLC). Permission for this study was granted by the Ethics Committee of The Fifth People’s Hospital of Suzhou (No. 2016010), and the study was conducted according to the 2013 revision of the Declaration of Helsinki guidelines. Informed consent was obtained from all patients involved in this study.
Cell lines and culture
Human NSCLC cell lines A549 and H1299 were obtained from the American Type Culture Collection and cultured in RPMI-1640 (GE Healthcare HyClone, USA) supplemented with 10% FBS (GE Healthcare HyClone, USA).
LGR5 transfection in A549 and H1299 cells
The full-length LGR5 cDNA was amplified by PCR, and the primer sequences were as follows: 5'-CCAACTTTGTGCCAACCGGTCGCCACCATGGACACCTCCCGGCTCGG TGTGCTC-3' and 5'-AATGCCAACTCTGAGCTTGAGACATGGGACAAATGCCACAGAGG-3'.
The LGR5 DNA fragment was cloned into the AgeI and NheI sites of the lentivirus vector GV341 (Gene Chem, China), which was then transfected into A549 and H1299 cells. Cells transfected with the empty vector were used as controls. Stable LGR5-expressing clones were selected by puromycin (Gene Chem, China).
Quantitative reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was isolated from cells and tissues by the TRIzol method. 1µg of total RNA was used for cDNA synthesis, and cDNA was prepared by using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, USA). Quantitative RT-PCR analysis was performed on StepOne Plus (ABI) using the Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, USA). The primer sequences for GAPDH were 5'-ATCATCCCTGCCTCTACTGG-3' and 5'-TTTCTAGACGGCAGGTCAGGT-3', and those for LGR5 were 5'-GCAAACCTACGTCTGGACAA-3' and 5'-TGATGCTGGAGCTGGTAAAG-3'. The primer sequences for NOTCH1, SMAD2, NANOG, and OCT4 were 5'-GAGGCGTGGCAGACTATGC-3' and 5'-CTTGTACTCCGTCAGCGTGA-3'; 5'-CCGACACACCGAGATCCTAAC-3' and 5'-GAGGTGGCGTTTCTGGAATATAA-3'; 5'-TTTGTGGGCCTGAAGAAAACT-3' and 5'-AGGGCTGTCCTGAATAAGCAG-3'; and 5'-CTTGAATCCCGAATGGAAAGGG-3' and 5'-GTGTATATCCCAGGGTGATCCTC-3', respectively. The reaction system was as follows: 10 µL of Power SYBR Green PCR Master Mix, 1 µL of primer, 8 µL of H2O, and 1 µL of template cDNA. The mRNA expression was calculated using the 2−△Ct method.
Flow cytometry analysis
The expression of LGR5 in transfected A549/H1299 cells was measured using the rat anti-human LGR5 antibody (Miltenyi Biotec, Germany) according to the manufacturer’s instructions. The cells were suspended at a density of 1×106 cells/mL in Hank’s balanced salt solution (HBSS) with 1% FCS, incubated with the antibody (10 µL) for 30 min, and then washed once by HBSS with 1% FCS. Flow cytometry was performed on an FC 500 analyzer (Beckman Coulter). Data were analyzed by FlowJo (San Carlos, CA, USA).
Cell proliferation assay
Transfected cells were seeded in 96-well plates at a density of 3×103 cells per well, and cell proliferation was examined after 0, 24, 48, 72, and 96 h. At each time point, 10 µL of CCK8 (Vazyme, China) solution was added to each well and incubated for 2 h at 37 °C. A microplate reader (Thermo Fisher Scientific, USA) was used to detect the optical density at a wavelength of 450 nm.
Clone formation assay
Transfected cells were seeded in 6-well plates at a density of 2×102 cells per well. The medium was replaced every 3 days, and the cells were cultured for 12 days. The clones were fixed with a 4% paraformaldehyde solution for 15 min and stained with 0.5% crystal violet for 5 min (Sigma). A clone was defined as >50 cells. Clone formation rate as a percentage was determined according to the following formula: clone formation = (the number of clones/the number of seeded cells) ×100%. The detection was performed 3 times.
Wound-healing assay
Transfected cells were seeded at a density of 2×104 cells per well in 24-well plates and grown to full confluence overnight. The cells were scratched by a plastic tip, washed with PBS 3 times to remove the detached cells, and maintained in a 0.5% FBS-containing RPMI-1640. The cells were incubated at 37 °C in a 5% CO2 incubator for 24 h. Images of migrating cells were taken at 0, 12, and 24 h after scratching. The widths of the scratches were measured by Image-Pro Plus. The wound-healing assay was performed at least 3 times.
Invasion assays
The upper portion of the Transwell inserts with an 8 µm pore size was coated with 30 µL of Matrigel diluted 1:4 in serum-free RPMI-1640 and incubated at 37 °C for 4 h. A total of 2×104 cells were plated on coated inserts in serum-free medium, and the coated inserts were placed in a 24-well plate with 500 µL RPMI-1640 containing 20% FBS in the bottom chamber. Noninvaded cells and gel were removed from the upper chamber after 24 h incubation at 37 °C in a 5% CO2 incubator. The cells on the bottom of the membrane were fixed with anhydrous ethanol and then stained with crystal violet (Sigma). The number of invasive cells was counted in 5 random fields per filter at 100-fold magnification. This experiment was performed in triplicate.
Western blot
Cells were washed 3 times in cold PBS and lysed in RIPA buffer. The proteins were separated on 10% SDS-PAGE gels and then blotted onto PVDF membranes. The membrane was blocked with 5% milk and incubated with anti-NOTCH1 antibody (Abcam, UK) overnight and then incubated with HRP-conjugated secondary antibody (Abcam, UK) for 2 h. The proteins were detected using an ECL reagent kit (Thermo Fisher Scientific, USA).
Statistical analysis
Statistical analysis was performed using the SPSS 22.0 software (SPSS Inc., Chicago, USA). T-test, one-way ANOVA, and variance analyses were used as the statistical methods for the analysis of significant diferences, and a P value of <0.05 was considered statistically significant.
Results
LGR5 expression was markedly higher in NSCLC tissues than in matched nontumor lung tissues
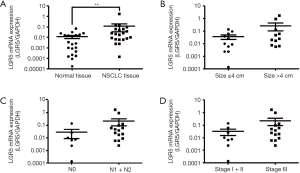
To explore the expression pattern of LGR5 and its clinical significance in NSCLC, we detected its expression by quantitative RT-PCR in NSCLC patients. LGR5 expression was significantly higher in NSCLC tissues than in adjacent nontumor lung tissues (P=0.0071) (Figure 1A). LGR5 expression showed no significant differences based on tumor size, lymph node status or TNM stage (P=0.2934, 0.1210 and 0.2601, respectively) (Figure 1B,C,D).
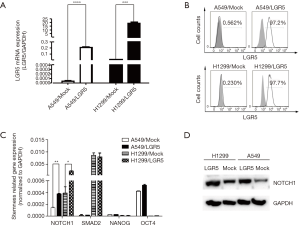
The expression of LGR5 was detected in transfected cells
Stable LGR5-overexpressing cells (A549/LGR5 and H1299/LGR5) were established to investigate the role of LGR5 in tumor promotion in NSCLC. Cells transfected with an empty vector were used as controls (A549/Mock and H1299/Mock). Quantitative RT-PCR and flow cytometry were used to examine the expression of LGR5 in the transfected cells. A higher expression of LGR5 mRNA was detected in A549/LGR5 and H1299/LGR5 cells compared with that in the controls (P<0.0001, P=0.0006, respectively) (Figure 2A). The expression of LGR5 in these cell lines was then assessed by flow cytometry, and the frequency of positive cells reached 97.2% and 97.7% in A549/LGR5 and H1299/LGR5 cells, respectively (Figure 2B). The results showed that we established high-LGR5-expression cell lines. When detecting the stemness-related gene expression in the transfected cells, we only found that the expression of NOTCH1 increased in both A549/LGR5 and H1299/LGR5 cells compared to that in A549/Mock and H1299/Mock cells (P=0.0036, P=0.0192, respectively) (Figure 2C,D).
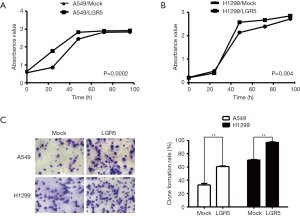
LGR5 promoted the proliferation and clone formation ability of NSCLC cells
To identify the biological function of LGR5 in NSCLC cell growth, we evaluated the proliferation and clone formation abilities of A549/Mock, A549/LGR5, H1299/ Mock, and H1299/LGR5 cells. The proliferation of these cells was detected at 0, 24, 48, 72, and 96 h by CCK-8 assay. The results showed that compared with the mock cells, LGR5-overexpressing cells had a higher rate of proliferation (P=0.0002, P=0.004) (Figure 3A,B). Next, we used the clone formation assay to test the cell proliferation ability further. The investigation showed that overexpression of LGR5 promoted clone formation (P=0.0059, P=0.0018) (Figure 3C).
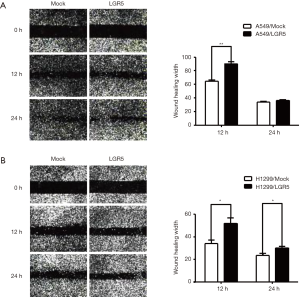
LGR5 enhanced the migration of NSCLC cells
As previously reported, LGR5 can promote the invasion and migration of colorectal cancer (16), esophageal squamous cell carcinoma (20) and gastric cancer (21). To further confirm the role of LGR5 in NSCLC cell migration, we performed a wound-healing assay with LGR5-transfected and Mock-transfected NSCLC cells. LGR5 enhanced the migration of A549 cells after 12 h (P=0.0029) (Figure 4A) and H1299 cells after 12 and 24 h (P=0.0387, P=0.0414, respectively) (Figure 4B).
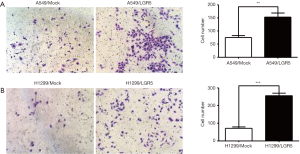
LGR5 increased the invasion ability of NSCLC cells
A Transwell invasion assay was used to determine the number of invasive cells to compare the invasion ability of LGR5-transfected NSCLC cells with that of the Mock-transfected control cells. The results showed that both the LGR5-transfected A549 cells (Figure 5A) and LGR5-transfected H1299 cells (Figure 5B) had increased invasion ability compared to the control cells.
Discussion
Lung cancer, which is the leading cause of cancer-related death, is a hazard to human health worldwide. The current 5-year relative survival rate of lung cancer is 18% (22). Little improvement has been made in the survival of lung cancer compared with that of most other cancers. Finding useful therapeutic targets for NSCLC has significant clinical value. Previous studies have shown that LGR5 is a stem cell marker in CRC and can promote tumor progression in various cancers, including CRC (17), ovarian cancer (23), and cervical cancer (24). Although many studies have found that LGR5 was significantly increased in NSCLC tissues and was associated with the poor prognosis of NSCLC (19,25), the mechanism of LGR5 in modulating the biological functions of NSCLC cells remains unknown.
In the present study, we confirmed that LGR5 expression was markedly higher in NSCLC tissues than in matched nontumor lung tissues. However, the LGR5 expression only showed a trend associated with larger tumor size, lymph node metastasis, and higher TNM stage because of the small sample size. These results suggest that LGR5 plays an essential role in NSCLC development.
Using LGR5-overexpressing NSCLC cells, this study further investigated the molecular function of LGR5 in NSCLC. The results revealed that the proliferation ability of LGR5-overexpressing cells was significantly higher than that of the control groups. To further explore the role of LGR5 in cell proliferation and population dependence, we applied a clone formation assay to test the clone formation rates. The clone formation rates of LGR5-overexpressing cells were significantly higher than those of the control groups. These results demonstrated that the upregulated expression of LGR5 in NSCLC could promote proliferation and clone formation, which may lead to stronger tumorigenicity of tumor cells. Using a wound-healing assay and a Transwell assay, we found that the migration and invasion abilities of LGR5-overexpressing cells were higher than those in the control groups. These results suggest that an upregulated LGR5 in NSCLC may promote metastasis and invasion. To demonstrate whether LGR5 expression is associated with stemness in NSCLC, we detected the expression of the stemness-related gene in LGR5-overexpressing cells. Only the expression of NOTCH1 was increased in both A549/LGR5 and H1299/LGR5 cells compared to that in A549/Mock and H1299/Mock cells. The NOTCH signaling pathway participates in regulating cell proliferation and differentiation (26,27). A previous study showed that LGR5 could promote ovarian cancer progression via NOTCH1 signaling (23) and that the NOTCH signaling pathway could control the proliferation and differentiation of Lgr5+ cells (28). These results suggest that LGR5 could also promote NSCLC progression via the NOTCH signaling pathway.
Conclusions
In summary, LGR5 expression was higher in NSCLC patients, was closely associated with tumorigenicity, metastasis, and invasion in NSCLC cells and could increase NOTCH1 expression. The results suggested that the function of LGR5 in promoting NSCLC progression may be correlated with NOTCH1. Our study explored the function of LGR5 in the biological behavior of NSCLC for the first time. These results imply that LGR5 might be a new target for gene-targeted therapies for NSCLC. Further studies are needed to explore the molecular mechanisms and signaling pathways involved in regulating LGR5 dysregulation in NSCLC.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.01.24). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all patients. The study was approved by the Ethics Committee of The Fifth People’s Hospital of Suzhou (No. 2016010).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Hamra GB, Guha N, Cohen A, et al. Outdoor particulate matter exposure and lung cancer: a systematic review and meta-analysis. Environ Health Perspect 2014;122:906-11. [Crossref] [PubMed]
- Straif K, Cohen A, Samet J. Air Pollution and Cancer. Lyon: IARC Press; 2013.
- Li S, Wang R, Zhang M, et al. Proteomic analysis of non-small cell lung cancer tissue interstitial fluids. World J Surg Oncol 2013;11:173. [Crossref] [PubMed]
- Reck M, Heigener DF, Mok T, et al. Management of non-small-cell lung cancer: recent developments. Lancet 2013;382:709-19. [Crossref] [PubMed]
- Dalerba P, Clarke MF. Cancer Stem Cells and Tumor Metastasis: First Steps into Uncharted Territory. Cell Stem Cell 2007;1:241-2. [Crossref] [PubMed]
- Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea--a paradigm shift. Cancer Res 2006;66:1883-90; discussion 1895-6.
- Sullivan JP, Minna JD, Shay JW. Evidence for self-renewing lung cancer stem cells and their implications in tumor initiation, progression, and targeted therapy. Cancer Metastasis Rev 2010;29:61-72. [Crossref] [PubMed]
- Bakhshinyan D, Adile AA, Qazi MA, et al. Introduction to Cancer Stem Cells: Past, Present, and Future. Methods Mol Biol 2018;1692:1-16. [Crossref] [PubMed]
- MacDonagh L, Gray SG, Breen E, et al. Lung cancer stem cells: The root of resistance. Cancer Lett 2016;372:147-56. [Crossref] [PubMed]
- Lundin A, Driscoll B. Lung cancer stem cells: progress and prospects. Cancer Lett 2013;338:89-93. [Crossref] [PubMed]
- Kemper K, Prasetyanti PR, De Lau W, et al. Monoclonal antibodies against Lgr5 identify human colorectal cancer stem cells. Stem Cells 2012;30:2378-86. [Crossref] [PubMed]
- de Lau W, Peng WC, Gros P, et al. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev 2014;28:305-16. [Crossref] [PubMed]
- Planas-Paz L, Orsini V, Boulter L, et al. The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size. Nat Cell Biol 2016;18:467-79. [Crossref] [PubMed]
- Merlos-Suárez A, Barriga FM, Jung P, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 2011;8:511-24. [Crossref] [PubMed]
- Uchida H, Yamazaki K, Fukuma M, et al. Overexpression of leucine-rich repeat-containing G protein-coupled receptor 5 in colorectal cancer. Cancer Sci 2010;101:1731-7. [Crossref] [PubMed]
- de Sousa e Melo F. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 2017;543:676-80. [Crossref] [PubMed]
- Schepers AG, Snippert HJ, Stange DE, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 2012;337:730-5. [Crossref] [PubMed]
- Gao F, Zhou B, Xu JC, et al. The role of LGR5 and ALDH1A1 in non-small cell lung cancer: Cancer progression and prognosis. Biochem Biophys Res Commun 2015;462:91-8. [Crossref] [PubMed]
- Lv Z, Yu JJ, Zhang WJ, et al. Expression and functional regulation of stemness gene Lgr5 in esophageal squamous cell carcinoma. Oncotarget 2017;8:26492-504. [PubMed]
- Wang B, Chen Q, Cao Y, et al. LGR5 Is a Gastric Cancer Stem Cell Marker Associated with Stemness and the EMT Signature Genes NANOG, NANOGP8, PRRX1, TWIST1, and BMI1. PloS One 2016;11:e0168904. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Liu W, Zhang J, Gan X, et al. LGR5 promotes epithelial ovarian cancer proliferation, metastasis, and epithelial-mesenchymal transition through the Notch1 signaling pathway. Cancer Med 2018; [Epub ahead of print]. [PubMed]
- Cao HZ, Liu XF, Yang WT, et al. LGR5 promotes cancer stem cell traits and chemoresistance in cervical cancer. Cell Death Dis 2017;8:e3039. [Crossref] [PubMed]
- Ryuge S, Sato Y, Jiang SX, et al. The clinicopathological significance of Lgr5 expression in lung adenocarcinoma. Lung Cancer 2013;82:143-8. [Crossref] [PubMed]
- Fre S, Huyghe M, Mourikis P, et al. Notch signals control the fate of immature progenitor cells in the intestine. Nature 2005;435:964-8. [Crossref] [PubMed]
- MacGrogan D, D’Amato G, Travisano S, et al. Sequential Ligand-Dependent Notch Signaling Activation Regulates Valve Primordium Formation and Morphogenesis. Circ Res 2016;118:1480-97. [Crossref] [PubMed]
- Dai Q, Duan C, Ren W, et al. Notch Signaling Regulates Lgr5(+) Olfactory Epithelium Progenitor/Stem Cell Turnover and Mediates Recovery of Lesioned Olfactory Epithelium in Mouse Model. Stem Cells 2018;36:1259-72. [Crossref] [PubMed]


