
Role of PRMT5 in bladder cancer: a comprehensive study
Introduction
Bladder cancer (BCa) is a very aggressive malignant tumor in the urinary system and has become the eighth most common cancer worldwide (1). The incidence and mortality rate of BCa have been clearly shown to increase with age (2). A high recurrence rate is a significant feature of BCa (3), but an early diagnosis and intervention for BCa patients can effectively improve their survival rates. Currently, the diagnosis of BCa depends on hematuria symptoms, cystoscopy, and urine abscission cytology (4). Since these traditional techniques are usually used to detect high-grade cancers, it is necessary to study the molecular mechanisms of, and discover the novel biomarkers for, the prognosis of BCa (5).
Protein arginine methyltransferase 5 (PRMT5) belongs to the methyltransferase family (6), and is involved in a wide range of cellular processes, such as the biosynthesis of organelles and transcriptional regulation (7). PRMT5 can catalyze the methylation of arginine in multiple target proteins, including histones, transcriptional elongation factors, and the tumor suppressor p53. Arginine methylation is an essential posttranscriptional protein modification that plays a crucial role in cell function. Methylation of H3 arginine 8 (H3R8) and H4 arginine 3 (H4R3) can induce the silencing of tumor suppressor genes, such as the suppressor of tumorigenicity 7 (ST7) and nonmetastatic 23 (NM23) (8). Abnormal expression of PRMT5 has been reported in many kinds of tumors, such as BCa (9,10), hepatocellular carcinoma (11), cervical cancer (12), breast cancer (13), lung cancer (14), glioma (15), and prostate cancer (16). Recently, one study reported that circulating PRMT5 (circPRMT5) was up-regulated in urothelial carcinoma of the bladder (UCB) tissues when compared with non-cancerous matched tissues, and its overexpression was positively associated with an advanced T and N status. Interestingly, this study also found that circPRMT5 was upregulated in both urine exosomes and serum from UCB patients, and both could also be used in predicting metastasis in UCB patients (9). Despite these studies, the role of PRMT5 in tumors is still not very clear.
In this work, the expression of PRMT5 in BCa and its clinicopathological significance were evaluated with an analysis of The Cancer Genome Atlas (TCGA) database and RT-qPCR data. To further confirm the effect of PRMT5 on the proliferation and migration of BCa cells, we constructed a PRMT5 knockdown model by lentivirus-mediated shRNA infection of SW780 cells. The results showed that the expression level of PRMT5 was upregulated in BCa tissues and cells.
Methods
Data extraction from the TCGA database
The clinicopathological data of PRMT5 in BCa were downloaded from the TCGA database (https://cancergenome.nih.gov/). Statistical analysis of TCGA data was performed by SPSS 22.0 software (IBM), and the expression levels of PRMT5 are presented as the mean ± SD. The expression levels of PRMT5 with different clinicopathological parameters were evaluated by independent sample t-tests. All the BCa samples were divided into two groups based on the mean value of the PRMT5 expression. Overall survival (OS) and recurrence-free survival (RFS) curves were utilized to measure the influence of PRMT5 on BCa prognosis.
Cell culture
Human bladder transitional cell carcinoma (SW780), human normal urothelial cells (SV-40-immortalized human uroepithelial cells, SV-HUC-1), and human embryonic kidney cells (293T) were purchased from the American Type Culture Collection (ATCC). The SW780 and 293T cells were cultured in DMEM (Invitrogen) supplemented with a 10% fetal bovine serum (FBS) and 1% antibiotics (100 U/mL penicillin and 100 µg/mL streptomycin sulfate). The SV-HUC-1 cells were maintained in F12K medium (Invitrogen, Carlsbad, CA, USA) with the same concentrations of FBS and antibiotics. All cells were cultured in incubators at 37 °C with a 5% CO2 atmosphere.
Construction of stable cell lines
A lentivirus vector suppressing PRMT5 gene expression was designed and synthesized by Syngen Tech (Beijing, China). The shRNA-PRMT5 and control shRNA were designed according to the PRMT5 gene sequence. The shRNA-PRMT5 sequence was 5'-CAGGAAGAGGGCCTATTTCCC-3'; the control shRNA sequence without significant homology was 5'-TAATTGTCAAATCAGAGTGCTT-3'. Lentivirus was produced by transfection of the plasmids into 293T cells using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol. After transfection for 48 or 72 h, lentivirus was harvested and passed through a 0.45-µm pore size filter (Millipore).
Next, the virus particles were concentrated and stored at −80 °C. To construct the stable cell line, SW780 was infected by the virus with polybrene (8 µg/mL, HanBio Biotechnology, Pudong, China). At 48 h postinfection, puromycin (1 µg/mL, Sigma–Aldrich, St. Louis, MO, USA) was used to screen the stable cell line.
RNA extraction and real-time quantitative PCR
Total RNA was extracted with the TRIzol reagent (Invitrogen, Grand Island, NY, USA) according to the manufacturer’s protocols. The concentration of the total RNA was measured by a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA). Total RNA was reverse transcribed into cDNA using a Revertra Ace qPCR RT Kit (Toyobo, Osaka, Japan). Real-time quantitative polymerase chain reactions (RT-qPCRs) were performed on the ABI PRISM 7000 Fluorescent Quantitative PCR system (Applied Biosystems, USA) with SYBR Green Premix (TaKaRa Bio Inc., Shiga, Japan).
All samples were normalized to GAPDH. The 2−ΔΔCt value was used to calculate the gene expression levels. The primer sequences used are as follows: PRMT5 primers: forward, 5'-TGTCTTCCATCCGCGTTTCA-3'; and reverse, 5'-AGCAGGAAAGCTGGAAGACC-3'. GAPDH primers: forward, 5'-AATCCCATCACCATCTTCCAG-3'; and reverse, 5'-TTCACACCCATGACGAACAT-3'.
Western blot assay
Cells were washed in PBS and lysed in RIPA buffer (50 mM Tris-HCl pH 7.2, 150 mM NaCl, 1% NP40, 0.1% SDS, 0.5% DOC, 1 mM PMSF, 25 mM MgCl2, and supplemented with a phosphatase inhibitor cocktail). The protein concentration was determined using the BCA protein assay. Equal amounts of whole protein extract were electrophoresed onto SDS–polyacrylamide gels and then transferred to PVDF membranes (Millipore, Billerica, MA, USA). Samples were blocked in a 5% dry milk solution and incubated overnight with the primary antibodies (Abcam, Cambridge, MA, USA). Then, the samples were incubated with horseradish peroxidase-conjugated secondary antibody (Amersham, Piscataway, NJ, USA) and immunoblots were developed with SuperSignal chemiluminescence reagents (Pierce Chemical Co.). The protein bands were quantified using Image J analysis software (National Institutes of Health, USA). Histograms were generated by normalizing the amount of each protein to the GAPDH level detected in the same extracted sample. Each experiment was repeated three times.
Cell proliferation assay
The Cell Counting Kit-8 assay (CCK-8; TransGen, Beijing, China) was used to assay cell proliferation. shRNA-PRMT5 SW780 cells or control shRNA SW780 cells were cultured in 96-well plates for several hours (0, 24, 48, or 72 h). In total, 10 µL of CCK-8 reagent was added to each well, and the cells were incubated for 2 h. A microplate reader (Bio-Rad, Hercules, CA, USA) was used to measure the absorbance at 450 nm.
Cell migration assay
Wound-healing and Transwell assays were used to measure cell migration ability. For the wound-healing assay, cells were cultured in 6-well plates with DMEM supplemented with 1% FBS and 1% antibiotics. A clear line was generated by a sterile pipette tip, and the cells were washed twice with PBS. At 0 or 24 h, the wound areas were photographed using a digital camera system, and the cell migration distance was calculated by the software program HMIAS-2000.
Transwell inserts with 8.0 µm pores (Dow Corning Corp., Midland, MI, USA) were used for Transwell assays. In total, 4×104 SW780 cells were cultured in 200 µL serum-free medium for 24 h and then placed in the uncoated dishes. The lower chambers were filled with 500 µL of medium containing 10% FBS. After 24 h, the migrated cells under the surface of the filter membrane were washed twice with 1× PBS, fixed with 4% paraformaldehyde, stained with 0.1% crystal violet (Sigma-Aldrich), and washed three times. Then, the invaded cells were photographed using an inverted microscope.
Statistical analysis
SPSS 22.0 software (IBM) was used to perform the statistical analyses. All data is presented as the mean ± SD. Independent samples t-tests and ANOVA were used to compare the differences. Here, P<0.05 was considered statistically significant (*P<0.05; **P<0.01).
Results
PRMT5 was highly expressed in BCa
In TCGA database, 407 BCa tissue samples and 19 adjacent normal tissues were evaluated to assess the expression levels of PRMT5 in tumor tissues [Figure 1A (P<0.05) and Figure 1B (P<0.01)]. We also measured the expression levels of PRMT5 by RT-qPCR in SW780 cells and SV-HUC-1 cells, coinciding with the results of TCGA database analysis [Figure 1C (P<0.05)]. These results suggested that PRMT5 was significantly upregulated in BCa cells.

PRMT5 knockdown model constructed by lentivirus-mediated shRNA infection of SW780 cells
To further investigate the biological role of PRMT5 in BCa cells, stable cell lines were constructed with SW780 cells. As shown in Figure 2A, GFP expression confirmed the successful packaging of the lentivirus in the 293T cells. The produced viral particles were used to infect the SW780 cells, and PRMT5 shRNA or control shRNA was stably expressed. The GFP expression in the infected cells was screened with puromycin (Figure 2B). The knockdown efficiency was tested by RT-qPCR and a western blot assay. The results showed that the lentivirus-mediated shRNA had a specific effect on PRMT5 in SW780 cells and that the knockdown efficiency was approximately 70% [Figure 2C (P<0.01)]. The results of the western blot showed that the shRNA-PRMT5 effectively reduced PRMT5 protein expression in SW780 cells (Figure 2D).
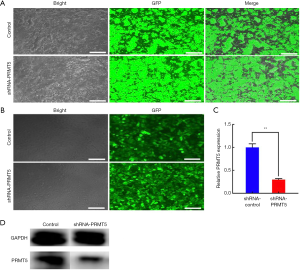
The effect of PRMT5 knockdown on cell proliferation and migration
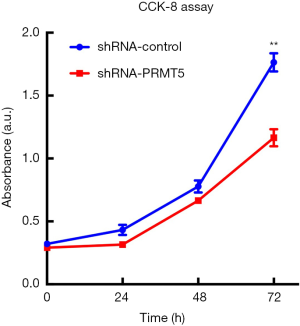
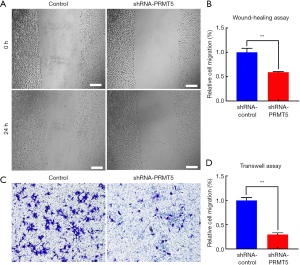
The CCK-8 assay was used to verify the effects of the PRMT5 knockdown on growth and proliferation of the BCa cells. The results showed that downregulation of PRMT5 could suppress the proliferation ability of SW780 cells within 72 h [Figure 3 (P<0.01)]. Wound-healing and Transwell assays were performed to assess the effect of PRMT5 on cell migration. In the wound-healing assay, cell migration ability was evaluated by calculating cell migration distances after a 24 h incubation. The results showed that SW780 cell migration was significantly inhibited due to PRMT5 knockdown [Figure 4A,B (P<0.01)]. The results of the Transwell assay were similar to the results of the scratch assay. As shown in Figure 4C,D (P<0.01), PRMT5 knockdown sharply reduced the migration of SW780 cells. These results indicated that downregulation of PRMT5 can efficiently inhibit cell migration.
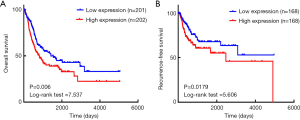
The relationship between clinicopathological features and PRMT5 expression in BCa
Information on 403 patients from TCGA database was divided into two groups according to the average expression of PRMT5. In Kaplan-Meier survival analyses, the log-rank test was used to evaluate the effects of elevated PRMT5 expression on OS and RFS. According to the patient data downloaded from TCGA database, PRMT5 expression was ranked, patients with missing survival data were eliminated, and the median was selected as the cut-off value. Due to the lack of follow-up data, the number of patients in the OS and RFS groups was different. As shown in Figure 5, the expression level of PRMT5 was negatively correlated with OS (P=0.006, log-rank test =7.537) and RFS (P=0.0179, Log-rank test =5.606). However, other clinicopathological features, such as tumor stage, neoplasm histologic grade, the presence of lymph-vascular invasion, pathologic stage, pathologic tumor, pathologic node, pathologic metastases, and tumor location, were not correlated with the expression level of PRMT5 (data not shown).
Discussion
In this work, we investigated whether PRMT5 functions as an oncogene that plays a vital role in BCa. The expression level of PRMT5 in BCa was examined by TCGA database analysis and RT-qPCR, and the biological function of PRMT5 was further explored by the construction of a PRMT5 knockdown model. The expression level of PRMT5 in BCa tissues and cells was higher than that in adjacent normal tissues and normal urinary epithelial cells (SV-HUC-1). PRMT5 knockdown could significantly inhibit the proliferation and migration of BCa cells. These results demonstrated that PRMT5 could be a novel biomarker for the prognosis of BCa.
Through TCGA database analysis, RT-qPCR and in vitro experiments, we comprehensively studied the expression and function of PRMT5 and evaluated the relationship between PRMT5 and BCa clinical prognosis. A large number of samples in the database could compensate for the limitations of our in vitro experiments. The database analysis also helped to preliminarily explore the research direction, such as the high expression of PRMT5 in BCa and the association between PRMT5 and clinicopathological features. The RT-qPCR and in vitro experiments were used to verify the conclusions further, and their results were consistent with those of TCGA database analysis in our study. Therefore, a combination of various methods could effectively screen molecular targets for tumor diagnosis and treatment.
Currently, the main tasks of cancer research are to find new prognostic markers, to improve the early diagnosis rate and to provide novel molecular targets. PRMT5 is an important member of the PRMT enzyme family. Many studies have shown that PRMT enzymes are closely related to DNA repair, signal transduction, and gene transcription. As the most important symmetrical arginine methyltransferase, PRMT5 is widely expressed and regulates many cellular functions, including differentiation, proliferation, growth, and apoptosis.
PRMT5 expression is abnormal during carcinogenesis in such cancers as gastric cancer, hepatocellular carcinoma, cervical cancer, breast cancer, lung cancer, glioma, and prostate cancer (9-19). Silencing tumor suppressor genes, such as SMAD7, RBL2, and NM23, ST7 (8,20,21), is one of the carcinogenic mechanisms of PRMT5. However, to understand the complex and multifactorial processes involved, such as tumor occurrence, growth and metastasis, mechanisms and pathways are needed for further study. As a potential biomarker for cancer prognosis, a study of the molecular mechanism of PRMT5 is critical. Therefore, our subsequent research will focus on the mechanism of PRMT5 in BCa and provide a theoretical basis for PRMT5 to become an effective target for cancer therapy.
Conclusions
In summary, PRMT5 was highly expressed in BCa, and silencing of PRMT5 expression could significantly inhibit cell proliferation and migration. Based on the Kaplan-Meier survival analyses, the expression level of PRMT5 was negatively correlated with the prognosis of BCa. These results suggested that PRMT5 plays a crucial role in the growth of BCa and could be used as a novel tumor biomarker for the prognosis and treatment of BCa.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.03.05). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Antoni S, Ferlay J, Soerjomataram I, et al. Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur Urol 2017;71:96-108. [Crossref] [PubMed]
- Tan WS, Rodney S, Lamb B, et al. Management of non-muscle invasive bladder cancer: A comprehensive analysis of guidelines from the United States, Europe and Asia. Cancer Treat Rev 2016;47:22-31. [Crossref] [PubMed]
- Sanli O, Dobruch J, Knowles MA, et al. Bladder cancer. Nat Rev Dis Primers 2017;3:17022. [Crossref] [PubMed]
- Grivas PD, Melas M, Papavassiliou AG. The biological complexity of urothelial carcinoma: Insights into carcinogenesis, targets and biomarkers of response to therapeutic approaches. Semin Cancer Biol 2015;35:125-32. [Crossref] [PubMed]
- Kaniskan HU, Jin J. Recent progress in developing selective inhibitors of protein methyltransferases. Curr Opin Chem Biol 2017;39:100-8. [Crossref] [PubMed]
- Blanc RS, Richard S. Arginine Methylation: The Coming of Age. Mol Cell 2017;65:8-24. [Crossref] [PubMed]
- Pal S, Vishwanath SN, Erdjument-Bromage H, et al. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol 2004;24:9630-45. [Crossref] [PubMed]
- Chen X, Chen RX, Wei WS, et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin Cancer Res 2018;24:6319-30. [Crossref] [PubMed]
- Sipos A, Ivan J, Becsi B, et al. Myosin phosphatase and RhoA-activated kinase modulate arginine methylation by the regulation of protein arginine methyltransferase 5 in hepatocellular carcinoma cells. Sci Rep 2017;7:40590. [Crossref] [PubMed]
- Jeon JY, Lee JS, Park ER, et al. Protein arginine methyltransferase 5 is implicated in the aggressiveness of human hepatocellular carcinoma and controls the invasive activity of cancer cells. Oncol Rep 2018;40:536-44. [PubMed]
- Dong SH, Wang X, Tian SC, et al. Arginine methyltransferase inhibitor 1 exhibits antitumor effects against cervical cancer in vitro and in vivo. Pharmazie 2018;73:269-73. [PubMed]
- Chiang K, Zielinska AE, Shaaban AM, et al. PRMT5 Is a Critical Regulator of Breast Cancer Stem Cell Function via Histone Methylation and FOXP1 Expression. Cell Rep 2017;21:3498-513. [Crossref] [PubMed]
- Jing P, Zhao N, Ye M, et al. Protein arginine methyltransferase 5 promotes lung cancer metastasis via the epigenetic regulation of miR-99 family/FGFR3 signaling. Cancer Lett 2018;427:38-48. [Crossref] [PubMed]
- Lu YF, Cai XL, Li ZZ, et al. LncRNA SNHG16 Functions as an Oncogene by Sponging MiR-4518 and Up-Regulating PRMT5 Expression in Glioma. Cell Physiol Biochem 2018;45:1975-85. [Crossref] [PubMed]
- Deng X, Shao G, Zhang HT, et al. Protein arginine methyltransferase 5 functions as an epigenetic activator of the androgen receptor to promote prostate cancer cell growth. Oncogene 2017;36:1223-31. [Crossref] [PubMed]
- Liu X, Zhang J, Liu L, et al. Protein arginine methyltransferase 5-mediated epigenetic silencing of IRX1 contributes to tumorigenicity and metastasis of gastric cancer. Biochim Biophys Acta Mol Basis Dis 2018;1864:2835-44. [Crossref] [PubMed]
- Huang S, Chi Y, Qin Y, et al. CAPG enhances breast cancer metastasis by competing with PRMT5 to modulate STC-1 transcription. Theranostics 2018;8:2549-64. [Crossref] [PubMed]
- Li Z, Zhang J, Liu X, et al. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat Commun 2018;9:1572. [Crossref] [PubMed]
- Wang L, Pal S, Sif S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol Cell Biol 2008;28:6262-77. [Crossref] [PubMed]
- Pal S, Baiocchi RA, Byrd JC, et al. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J 2007;26:3558-69. [Crossref] [PubMed]


