
This article has an erratum available at: http://dx.doi.org/10.21037/tcr-2023-01 the article has been update on 2023-04-13 at here.
Long non-coding RNA SNHG17 promotes gastric cancer progression by inhibiting P15 and P16
Introduction
Gastric carcinoma remains one of the most prevalent tumors of digestive system, and the incidence of stomach cancer in China, Japan, Latin America, and Eastern Europe is relatively higher than in other countries. Approximately 72 million people die from this disease every year worldwide (1,2). Gastric cancer endangers human life, and radical surgery is the only potential curative approach. With the increase of early gastric cancer detection and the progress of comprehensive treatment, the prognosis of gastric cancer patients has greatly improved; however, the long-term survival of advanced gastric cancer patients remains poor (3). Gastric cancer development is a complex and multistage biological process that involves multiple oncogenes and tumor suppressors (4,5). The goal of genomic studies in gastric cancer is to find early diagnostic biomarkers and/or novel therapeutic targets.
Human genome analyses have indicated that 85% of DNA can be transcribed into RNA, but less than 2% of RNA can be translated into protein. The RNAs that cannot be translated into proteins are called non-coding RNAs (ncRNAs) (6). NcRNAs can be classified into short ncRNAs, mid-sized ncRNAs, and long ncRNAs (lncRNAs) on the basis of molecular length (7). LncRNAs are defined as being >200 bp in length, and >15,000 different types of lncRNAs have been identified by gene chip or high-throughput sequencing technologies (8). LncRNAs regulate cellular functions at the gene level by participating in genomic imprinting, chromatin packaging, differentiation, maintaining genomic integrity, and affecting embryo development (9,10). Abnormal lncRNA expression in tumor cells is usually associated with increased proliferation, invasion, and angiogenesis; however, other studies have shown that lncRNAs can also inhibit apoptosis and affects cell cycle progression in tumor cells (11-13).
Accumulating evidence has demonstrated that cancer involves a disorder of cell cycle regulatory mechanisms (14-16). There are three major protein families that regulate cell cycle progression: the cyclins, the cyclin dependent kinases (CDKs), and the CDK inhibitors (CKIs). Cyclins combine with CDKs to form a complex that drives cell cycle progression, and CKIs represses cyclin-CDK complexes (17). Currently, seven CKIs have been identified, and they are divided into two families: the INK4 and CIP/KIP families. The former includes P15, P16, P18 and P19, which competes with cyclin D1-, D2-, and D3-CDK4/6 complexes in the G1 phase of the cell cycle to block cell cycle progression. CIP/KIP family includes P21, P27, and P57, which bind to cyclin-CDK2/7/9/10 complexes, acting on the G0/G1, S, and G2/M phases (18-20). Recently, CKIs have been shown to acquire tumor-suppressive properties in certain environments (21), while lncRNAs have been shown to promote tumor cell proliferation and migration by decreasing the expression of CKIs (22-24).
The roles of some small nucleolar RNA host genes (SNHGs) in cancer have attracted much attention recently. SNHG5 suppresses gastric cancer progression (25), while SNHG6 is related to poor prognosis in stomach carcinoma and promotes cell proliferation (22). However, the roles of SNHG17 in stomach cancer remain to be investigated. In the present study, we first consulted The Cancer Genome Atlas (TCGA), and detected that SNHG17 expression was remarkably higher in the gastric cancer tissues than in normal stomach mucosae. We found that SNHG17 was overexpressed in stomach cancer tissues comparing to corresponding normal tissues, and that SNHG17 was associated with lymph node metastasis, pTNM stage, and lymphovascular invasion. Loss-of-function assays showed that SNHG17 inhibited P15 and P16 and upregulated CDK4, resulting in cell cycle arrest at the G0/G1 phase. Subsequently, SNHG17 promoted gastric cancer cell proliferation and migration. This preliminary study elucidated the roles and mechanisms of the lncRNA SNHG17 in gastric cancer. These results could provide a new biomarker or target for diagnosing or treating gastric cancer.
Methods
Human gastric cancer specimens
Patients with stomach cancer (n=80; median age: 59 years; range, 36–76 years) from January 2012 to December 2013 at the First Affiliated Hospital of Nanjing Medical University were randomly enrolled in this study. All these patients were pathologically diagnosed as gastric adenocarcinoma according to the American Joint Committee on Cancer criteria. All these patients had not received preoperative chemotherapy or radiotherapy. Samples of tumor tissues and corresponding noncancerous mucosae were collected immediately following resection and snap frozen in liquid nitrogen. All patients were followed up until March 2018, with a median follow-up of 48 months. Overall survival (OS) was defined as the interval between the dates of surgery and death. Distant metastases were not observed preoperatively in these patients. This study was approved by the Nanjing Medical University Institutional Review Board. All the patients provided the written informed consent for their information and samples to be stored in the hospital database and used for researches. This study was compliant with the Helsinki Declaration.
Cell culture
We applied the normal human stomach epithelial cell line GES-1 (CBTCCCAS, Shanghai, China) and the human gastric adenocarcinoma cell lines AGS, SGC7901, BGC803, and BGC823 (ATCC, Manassas, VA, USA) in this study, and these cell lines were cultured in DMEM and RPMI-1640 (Invitrogen, Carlsbad, CA, USA), respectively at 37 °C in a humidified atmosphere with 5% CO2, which was supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA), penicillin/streptomycin (1:100 dilution; Invitrogen), and 4 mM glutamine (Life Technologies, Gibco BRL).
Real-time quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR analyses were performed with reference to our previous report (26). The following primer pairs were applied in this study: SNHG17: 5'-TGCTTGTAAGGCAGGGTCTC-3' (sense) and 5'-ACAGCCACTGAAAGCATGTG-3' (antisense); P15: 5'-GGACTAGTGGAGAAGGTGCG-3' (sense) and 5'-GGGCGCTGCCCATCATCATG-3' (antisense); P16: 5'-ATGGAGCCTTCGGCTGAC-3' (sense) and 5'-GGCCTCCGACCGTAACTA-3' (antisense); P18: 5'-CGCGGATCCGCGCATG-3' (sense) and 5'-CCGCTCGAGCGGTCAC-3' (antisense); P19: 5'-GCGCCTGGTCACCAGGGC-3' (sense) and 5'-CACCTATAAGCCACAAAC-3' (antisense); and glyceraldehyde-3-phosphate dehydrogenase (GAPDH): 5'-GAAGAGAGAGACCCTCACGCT-3' (sense) and 5'-ACTGTGAGGAGGGGAGATTCA-3' (antisense). GAPDH served as the reference, and the observed gene expression levels were normalized to GAPDH. All assays were performed in triplicate.
Western blotting
Western blotting was used to analyze protein expression with reference to our previous report (26). The following antibodies were used: mouse monoclonal anti-P15, anti-P16, anti-CDK4, and rabbit monoclonal anti-GAPDH antibodies (Cell Signaling Technology, Danvers, MA, USA). Protein expression was quantified and normalized to GAPDH by densitometric analysis.
Transduction of SNHG17 siRNAs
Two SNHG17 siRNAs, siRNA-SNHG17#1 (AAACGAGCGTAGCTTCCTT) and siRNA-SNHG17#2 (GCAGTGTCTCGTCCTCTTT), were designed and synthesized. SNHG17 siRNA (siRNA-SNHG17) transduction was performed using riboFECTTM CP (RiboBio, Guangzhou, China) with reference to the manufacturer’s recommended protocol.
Cell proliferation assay
The Cell Counting Kit-8 assay (Sigma-Aldrich, St. Louis, MO, USA) was performed to assay cell proliferation. Results are plotted as mean ± standard error of three separate experiments for each condition.
Colony formation assay
The clonogenic potential of the cells was assessed by colony formation assays. Briefly, 500 single viable cells in RPMI-1640 or DMEM containing 10% FBS were plated in 6-well plates, and were incubated at 37 °C in an atmosphere of 5% CO2 for 14 d. Subsequently, the colonies formed were stained with 0.1% Crystal Violet Solution (Sigma-Aldrich), washed, and counted.
Transwell migration assay
According to the manufacturer’s protocol, cells (3×105 per well) were seeded in the upper chambers of 24-well Transwell inserts (8.0-µm pore size; Corning, NY, USA) with 200 µL RMPI-1640 or DMEM containing 1% fetal bovine serum and 0.2% bovine serum albumin. After culture, migrated cells were stained and counted.
Flow cytometry
Cells were stained with both FITC-Annexin V and propidium iodide (PI) using the FITC-Annexin V Apoptosis Detection Kit (BD Biosciences, Franklin Lakes, NJ, USA), and were analyzed by flow cytometry using a FACScan with CellQuest software (BD Biosciences). Cells were classified into viable cells, dead cells, early apoptotic cells, and apoptotic cells. The relative ratio of early apoptotic cells was compared with that of the controls for each experiment, and all samples were assayed at least in triplicate.
Tumorigenicity in vivo
All animal studies were conducted under the guidelines of the Nanjing Medical University Institutional Animal Care and Use Committee. A total of 16 4-week-old male BALB/c nude mice (Vitalriver, Beijing, China) were grouped randomly. For the analysis of tumorigenicity, 5×105 cells from AGS-si-NC, or AGS-si-SNHG17 stable cell lines were inoculated subcutaneously into the flanks of the mice. Tumor volumes were measured every 4 d and were calculated using the following formula: volume = (length × width2) ×0.5. On 40 d of inoculation, mice were euthanized, and tumor tissues were removed, weighed, and snap frozen in liquid nitrogen.
Statistical analysis
All statistical analyses were performed using SPSS software (version 17.0; SPSS Inc., Chicago, IL, USA). Data are presented as mean ± standard error unless indicated. All values in protein expression are representative of three independent experiments with similar results. Associations of lncRNA levels with various clinicopathological parameters were evaluated with Pearson’s χ2 test. Quantitative data between the control and treatment groups were analyzed by variance analysis. P less than 0.05 was defined as statistically significant.
Results
SNHG17 was overexpressed in human gastric cancer cell lines and tissues and associated with lymph node metastasis, pTNM stage, and lymphovascular invasion
SNHG17 expression was evaluated by RT-qPCR in GES-1, AGS, SGC7901, BGC803, and BGC823 cells. As indicated in Figure 1A, SNHG17 levels were higher in human gastric adenocarcinoma cell lines than in the human normal stomach epithelial cell line (P<0.01). We next analyzed TCGA database (www.cancergenome.nih.gov. version 2017-09-08). There were 375 cases of gastric cancer with SNHG17 value in tumor tissues and 32 of them had SNHG17 value in normal mucosae simultaneously, and analysis revealed that SNHG17 value in tumor tissues was higher than in normal mucosae (Figure 1B, P=4.85×10−10), indicating that SNHG17 was overexpressed in human stomach tumor tissues compared with noncancerous mucosae.
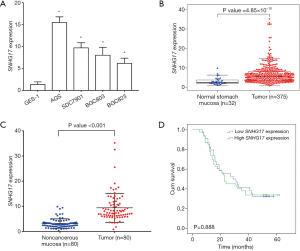
Furthermore, we examined SNHG17 expression in 80 human gastric cancer specimens, which indicated that SNHG17 levels were significantly higher in stomach tumor tissues than in corresponding normal mucosae (Figure 1C, P<0.0001). According to the median SNHG17 expression level, the 80 gastric cancer patients were divided into high group (above the median value, n=46) and low group (below the median value, n=34). Correlations between SNHG17 expression and clinicopathological characteristics were investigated. As indicated in Table 1, high SNHG17 expression in tumor tissue was associated with lymph node metastasis (P=0.0006), pTNM stage (P=0.0061), and lymphovascular invasion (P=0.0005), and was not significantly associated with gender, age, tumor location, tumor size, or OS (Figure 1D, P=0.888).
Table 1
| Clinicopathological features | N | SNHG17 expression in tumor tissue | P value | |
|---|---|---|---|---|
| High cases (%) | Low cases (%) | |||
| Age (years) | 0.1646 | |||
| <60 | 33 | 22 (66.7) | 11 (33.3) | |
| ≥60 | 47 | 24 (51.1) | 23 (48.9) | |
| Gender | 0.3130 | |||
| Male | 58 | 31 (53.4) | 27 (46.6) | |
| Female | 22 | 15 (68.2) | 7 (31.8) | |
| Lauren classification | 0.7818 | |||
| Intestinal | 48 | 27 (56.3) | 21 (43.7) | |
| Diffuse and mixed | 32 | 19 (59.4) | 13(40.6) | |
| Tumor size (cm) | 0.1408 | |||
| ≥3 | 54 | 28 (51.9) | 26 (48.1) | |
| <3 | 26 | 18 (69.2) | 8 (30.8) | |
| Tumor location | 0.9519 | |||
| Upper third | 28 | 16 (57.1) | 12 (42.9) | |
| Middle third | 20 | 11 (55.0) | 9 (45.0) | |
| Lower third | 32 | 19 (59.4) | 13 (40.6) | |
| Depth of tumor invasion | 0.4805 | |||
| Localized in subserosa | 27 | 17 (63.0) | 10 (37.0) | |
| Beyond subserosa | 53 | 29 (54.7) | 24 (45.3) | |
| Lymph node metastasis | 0.0006 | |||
| N0 | 25 | 7 (28.0) | 18 (72.0) | |
| N1–N3 | 55 | 38 (69.1) | 17 (30.9) | |
| pTNM stage | 0.0061 | |||
| I/II | 33 | 13 (39.4) | 20 (60.6) | |
| III/IV | 47 | 33 (70.2) | 14 (29.8) | |
| Lymphovascular invasion | 0.0005 | |||
| Absence | 52 | 22 (42.3) | 30 (57.7) | |
| Presence | 28 | 24 (85.7) | 4 (14.3) | |
Silencing SNHG17 inhibited gastric cancer cell proliferation and migration
To probe the effects of SNHG17 on cell behaviors, two SNHG17-targeted siRNAs, siRNA-SNHG17#1 and siRNA-SNHG17#2, were designed and synthesized. As shown in Figure 2A, both effectively silenced SNHG17 in AGS and SGC7901 cells (P<0.01).
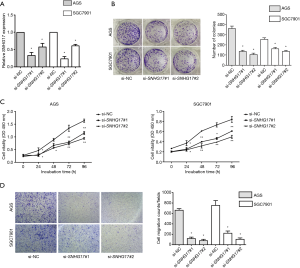
Colony formation assays indicated that colony numbers were significantly lower in SNHG17-silenced AGS and SGC7901 cells compared with controls (Figure 2B, P<0.01). Cell Counting Kit-8 assays also showed that silencing SNHG17 significantly reduced proliferation (Figure 2C, P<0.01). Transwell assays demonstrated that siRNA treatment significantly reduced the number of gastric tumor cells that migrated through Matrigel compared with the scrambled siRNA groups (Figure 2D, P<0.01). Together, these experimental and clinical results demonstrated that SNHG17 promoted gastric cancer progression.
Silencing SNHG17 resulted in G0/G1 cell cycle arrest and increased apoptosis in gastric cancer cell lines
Then, we investigated the effects of SNHG17 on gastric carcinoma cell cycle progression and apoptosis. As shown in Figure 3A, silencing SNHG17 with siRNA-SNHG17#1 or siRNA-SNHG17#2 resulted in a G0/G1 cell cycle arrest in AGS and SGC7901 cells (P<0.05). Additionally, flow cytometry indicated that silencing SNHG17 in AGS and SGC7901 cells significantly increased the percentage of apoptotic cells (Figure 3B, P<0.01). These findings suggested that SNHG17 regulates cell cycle progression and apoptosis, which may contribute to gastric cancer progression.
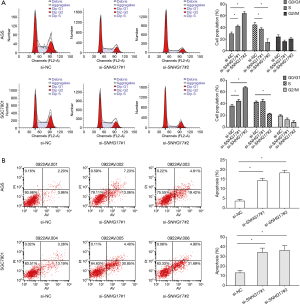
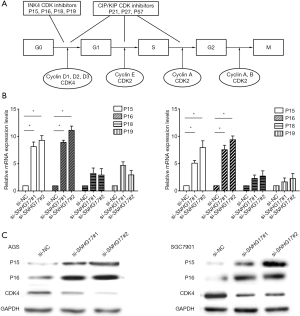
Silencing SNHG17 upregulated P15 and P16 expression and decreased CDK4 in gastric cancer cells
CKIs have been demonstrated to have functional diversity beyond cell cycle regulation, including regulation of cell proliferation, migration and apoptosis (17-19). To investigate the mechanism of how SNHG17 regulates cell cycle progression, we studied the effects of SNHG17 on the expression of INK4 family proteins and CDK4 (Figure 4A) in AGS and SGC7901 cells. As shown in Figure 4B, silencing SNHG17 significantly increased P15 and P16 mRNA expression (P<0.01), but did not change P18 and P19 levels. Western blotting analysis indicated that silencing SNHG17 upregulated P15 and P16 levels and decreased CDK4 (Figure 4C). Thus, it appears that SNHG17 inhibits P15 and P16, increasing proliferation and promoting gastric cancer progression.
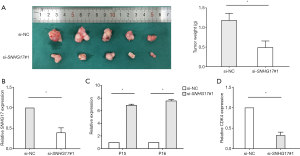
Silencing SNHG17 inhibited gastric cancer cell tumorigenicity in vivo
Human gastric cancer cell xenografts were established in nude mice to verify the role of SNHG17 in gastric cancer cell tumorigenicity. As shown in Figure 5, AGS cells transfected with si-SNHG17#1 (AGS-si-SNHG17#1) and AGS cells with control (AGS-si-NC) formed tumors after subcutaneously inoculation; however, tumor growth in the AGS-si-SNHG17#1 group was slower than in the AGS-si-NC group. Accordingly, reduced tumor weights were observed in the AGS-si-SNHG17#1 group compared with the control group after 40 d post-inoculation (Figure 5A; P<0.01). RT-qPCR assays of tumor tissues confirmed SNHG17 expression in the two groups (Figure 5B; P<0.05), and indicated that P15 and P16 expression were increased (Figure 5C; P<0.05), and CDK4 expression was decreased significantly (Figure 5D; P<0.05) in AGS-si-SNHG17#1 tumor compared with in AGS-si-NC tumors.
Discussion
Gastric cancer, which is an ongoing serious public health threat, represents the second leading cause of cancer mortality worldwide (26). Early detection, diagnosis, and effective prognostic indicators are necessary to improve the survival of gastric cancer patients. However, the lack of early diagnostic biomarkers, effective prognostic indicators, and therapeutic targets accounts for the current poor prognosis of gastric cancer patients. Thus, searching for novel molecular biomarkers is imperative.
There has been increasing interest in the epigenetic regulation of human cancers. Setting aside the Central Dogma, both miRNAs and lncRNAs are epigenetic regulators of human cancers (8,27). LncRNAs fold into various conformations that allow interactions with RNA, DNA and proteins to regulate nuclear, cytoplasmic and mitochondrial functions (28,29). LncRNAs regulate biological processes including transcription, chromatin dynamics, chromatin looping, histone modifications, telomere biology, protein complex assembly, RNA splicing, and translation (30). Many lncRNAs have been found to be dysregulated in cancers, and lncRNAs are thought to play indispensable roles in each of the hallmarks of cancer (31). The regulatory patterns of lncRNAs make cancer pathogenesis more intricate and complicated; however, the multi-functional and tissue-specific properties of lncRNAs provide new avenues for developing novel diagnostic, prognostic, and therapeutic biomarkers for cancers (11).
A myriad of evidence has indicated that lncRNAs are involved in gastric cancer development. Liu et al. (32) reported that upregulation of the lncRNA HOTAIR is related to larger tumor size, advanced pathological stage, extensive metastasis, and dismal OS in gastric cancer patients. The lncRNA ATB contributes to gastric cancer growth through a MiR-141-3p/TGFβ2 feedback loop (33), and FEZF1-AS1 is associated with adverse prognosis in stomach cancer and promotes tumorigenesis via activating the Wnt pathway (34). Currently, upregulated lncRNAs in cancer are thought to function as oncogenes the expedite the acquisition of the malignant cancer phenotypes, whereas the downregulated lncRNAs may possess tumor suppressive activities (11). For example, the lncRNA AC138128.1 is significantly repressed in gastric tumor tissues compared with adjacent noncancerous mucosae, and thus, is thought to be a potential biomarker for gastric cancer (35).
Non-coding multiple SNHGs have been suggested to play critical roles in regulating cell behaviors and carcinogenesis. Li et al. (22) reported that SNHG6 was overexpressed in human gastric cancer tissues and SNHG6 knockdown inhibited gastric cancer development by upregulating P21, which was dependent on the activation of the JNK pathway and suppression of enhancer of zeste homolog 2 (EZH2). SNHG1 expression is significantly higher in gastric tumor tissues than in adjacent normal mucosae, and SNHG1 accelerated the proliferation of gastric cancer cells and increased DNMT1 expression (36). SNHG14 overexpression contributes to gastric cancer development via targeting the miR-145/SOX9, which is involved in the PI3K/AKT/mTOR pathway (37). Conversely, SNHG5 is remarkably downregulated in stomach tumor tissues, as it suppresses gastric carcinoma cell proliferation, migration, and invasion by trapping MTA2 in the cytosol (25). Computationally, Zhao et al. (25) found that SNHG17 was upregulated in human gastric tumor tissues compared with normal mucosa tissues, and SNHG17 has been shown to be an unfavorable prognostic factor in colorectal cancer that promotes proliferation by epigenetically silencing P57 (38). Recently, Zhang et al. (39) reported that SNHG17 promotes gastric cancer progression by silencing P15 and P57.
Analysis of TCGA database revealed that SNHG17 was significantly overexpressed in stomach cancer tissues compared with noncancerous mucosae, and we verified this result in human gastric cancer specimens and cell lines. Furthermore, SNHG17 levels in tumor tissues were related to lymph node metastasis, pTNM stage, and lymphovascular invasion, but were not related to OS. Loss-of-function analyses indicated that SNHG17 promoted stomach carcinoma cell proliferation in vitro and in vivo, and that SNHG17 enhanced gastric cancer cell migration. Further assays demonstrated that SNHG17 inhibited P15 and P16, and enhances CDK4 expression, resulting in cell cycle arrest at G0/G1 phase, which was consistent with previous reports (39). Our study also showed that SNHG17 inhibited cell apoptosis. Thus, we inferred that SNHG17 promotes gastric cancer progression via inhibiting P15 and P16. However, how SNHG17 acts on P15 and P16 and whether other factors are involved in the process need to be further studied.
Currently, the concept of precision medicine and translational medicine has been well accepted (40), and certain dysregulated lncRNAs are promising candidate molecular biomarkers for cancer. Our preliminary findings highlight the role of SNHG17 in gastric cancer, and suggest that it may serve as a novel biomarker or therapeutic target for diagnosing and treating stomach carcinoma. However, further studies are needed to clarify the underlying mechanisms in detail.
Acknowledgments
The authors thank Dr. Ruji He, Second Affiliated Hospital of Soochow University, for his technical assistance.
Funding: This project was funded by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.04.14). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Nanjing Medical University Institutional Review Board. All the patients provided the written informed consent for their information and samples to be stored in the hospital database and used for researches. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Charalampakis N, Economopoulou P, Kotsantis I, et al. Medical management of gastric cancer: a 2017 update. Cancer Med 2018;7:123-33. [Crossref] [PubMed]
- Salati M, Di Emidio K, Tarantino V, et al. Second-line treatments: moving towards an opportunity to improve survival in advanced gastric cancer. ESMO Open 2017;2:e000206. [Crossref] [PubMed]
- Van Cutsem E, Sagaert X, Topal B, et al. Gastric cancer. Lancet 2016;388:2654-64. [Crossref] [PubMed]
- Nemtsova MV, Strelnikov VV, Tanas AS, et al. Implication of gastric cancer molecular genetic markers in surgical practice. Curr Genomics 2017;18:408-15. [Crossref] [PubMed]
- Sitarz R, Skierucha M, Mielko J, et al. Gastric cancer: epidemiology, prevention, classification, and treatment. Cancer Manag Res 2018;10:239-48. [Crossref] [PubMed]
- Awan HM, Shah A, Rashid F, et al. Primate-specific long non-coding RNAs and microRNAs. Genomics Proteomics Bioinformatics 2017;15:187-95. [Crossref] [PubMed]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011;12:861-74. [Crossref] [PubMed]
- Klinge CM. Non-coding RNAs: long non-coding RNAs and microRNAs in endocrine-related cancers. Endocr Relat Cancer 2018;25:R259-82. [Crossref] [PubMed]
- Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol 2013;10:925-33. [Crossref] [PubMed]
- Bhan A, Mandal SS. Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease. ChemMedChem 2014;9:1932-56. [Crossref] [PubMed]
- Zhao J, Liu Y, Huang G, et al. Long non-coding RNAs in gastric cancer: versatile mechanisms and potential for clinical translation. Am J Cancer Res 2015;5:907-27. [PubMed]
- Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta 2014;1839:1097-109. [Crossref] [PubMed]
- Zhao W, Geng D, Li S, et al. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med 2018;7:842-55. [Crossref] [PubMed]
- Mens MMJ, Ghanbari M. Cell cycle regulation of stem cells by microRNAs. Stem Cell Rev 2018;14:309-22. [Crossref] [PubMed]
- Boeynaems S, Tompa P, Van Den Bosch L. Phasing in on the cell cycle. Cell Div 2018;13:1. [Crossref] [PubMed]
- Saha P, Datta K. Multi-functional, multicompartmental hyaluronan-binding protein 1 (HABP1/p32/gC1qR): implication in cancer progression and metastasis. Oncotarget 2018;9:10784-807. [Crossref] [PubMed]
- Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development 2013;140:3079-93. [Crossref] [PubMed]
- Kumar A, Gopalswamy M, Wolf A, et al. Phosphorylation-induced unfolding regulates p19INK4d during the human cell cycle. Proc Natl Acad Sci U S A 2018;115:3344-9. [Crossref] [PubMed]
- Grimmler M, Wang Y, Mund T, et al. Cdk-inhibitory activity and stability of p27Kip1 are directly regulated by oncogenic tyrosine kinases. Cell 2007;128:269-80. [Crossref] [PubMed]
- Cheng CW, Leong KW, Ng YM, et al. The peptidyl-prolyl isomerase PIN1 relieves cyclin-dependent kinase 2 (CDK2) inhibition by the CDK inhibitor p27. J Biol Chem 2017;292:21431-41. [Crossref] [PubMed]
- Georgakilas AG, Martin OA, Bonner WM. p21: a two-faced genome guardian. Trends Mol Med 2017;23:310-9. [Crossref] [PubMed]
- Li Y, Li D, Zhao M, et al. Long noncoding RNA SNHG6 regulates p21 expression via activation of the JNK pathway and regulation of EZH2 in gastric cancer cells. Life Sci 2018;208:295-304. [Crossref] [PubMed]
- Su J, Zhang E, Han L, et al. Long noncoding RNA BLACAT1 indicates a poor prognosis of colorectal cancer and affects cell proliferation by epigenetically silencing of p15. Cell Death Dis 2017;8:e2665. [Crossref] [PubMed]
- Zhang F, Peng H. LncRNA-ANCR regulates the cell growth of osteosarcoma by interacting with EZH2 and affecting the expression of p21 and p27. J Orthop Surg Res 2017;12:103. [Crossref] [PubMed]
- Zhao L, Guo H, Zhou B, et al. Long non-coding RNA SNHG5 suppresses gastric cancer progression by trapping MTA2 in the cytosol. Oncogene 2016;35:5770-80. [Crossref] [PubMed]
- Wen J, Wang Y, Gao C, et al. Helicobacter pylori infection promotes Aquaporin 3 expression via the ROS-HIF-1α-AQP3-ROS loop in stomach mucosa: a potential novel mechanism for cancer pathogenesis. Oncogene 2018;37:3549-61. [Crossref] [PubMed]
- Liz J, Esteller M. LncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta 2016;1859:169-76. [Crossref] [PubMed]
- Delás MJ, Hannon GJ. LncRNAs in development and disease: from functions to mechanisms. Open Biol 2017;7:170121. [Crossref] [PubMed]
- De Paepe B, Lefever S, Mestdagh P. How long noncoding RNAs enforce their will on mitochondrial activity: regulation of mitochondrial respiration, reactive oxygen species production, apoptosis, and metabolic reprogramming in cancer. Curr Genet 2018;64:163-72. [Crossref] [PubMed]
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 2013;20:300-7. [Crossref] [PubMed]
- Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 2012;9:703-19. [Crossref] [PubMed]
- Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer 2014;13:92. [Crossref] [PubMed]
- Lei K, Liang X, Gao Y, et al. Lnc-ATB contributes to gastric cancer growth through a MiR-141-3p/TGFβ2 feedback loop. Biochem Biophys Res Commun 2017;484:514-21. [Crossref] [PubMed]
- Wu X, Zhang P, Zhu H, et al. Long noncoding RNA FEZF1-AS1 indicates a poor prognosis of gastric cancer and promotes tumorigenesis via activation of Wnt signaling pathway. Biomed Pharmacother 2017;96:1103-8. [Crossref] [PubMed]
- Chen X, Sun J, Song Y, et al. The novel long noncoding RNA AC138128.1 may be a predictive biomarker in gastric cancer. Med Oncol 2014;31:262. [Crossref] [PubMed]
- Hu Y, Ma Z, He Y, et al. LncRNA-SNHG1 contributes to gastric cancer cell proliferation by regulating DNMT1. Biochem Biophys Res Commun 2017;491:926-31. [Crossref] [PubMed]
- Liu Z, Yan Y, Cao S, et al. Long non-coding RNA SNHG14 contributes to gastric cancer development through targeting miR-145/SOX9 axis. J Cell Biochem 2018;119:6905-13. [Crossref] [PubMed]
- Ma Z, Gu S, Song M, et al. Long non-coding RNA SNHG17 is an unfavourable prognostic factor and promotes cell proliferation by epigenetically silencing P57 in colorectal cancer. Mol Biosyst 2017;13:2350-61. [Crossref] [PubMed]
- Zhang G, Xu Y, Wang S, et al. LncRNA SNHG17 promotes gastric cancer progression by epigenetically silencing of p15 and p57. J Cell Physiol 2019;234:5163-74. [Crossref] [PubMed]
- Wu JQ, Zhai J, Li CY, et al. Patient-derived xenograft in zebrafish embryos: a new platform for translational research in gastric cancer. J Exp Clin Cancer Res 2017;36:160. [Crossref] [PubMed]


