
Human cancer cells compensate the genes unfavorable for translation by N6-methyladenosine modification and enhance their translation efficiency
Introduction
RNA modification is omnipresent in all living organisms and cells. More than one hundred types of RNA modifications are known (1), of which the two most prevalent in the animal kingdom are A-to-I RNA editing and N6-methyladenosine (m6A). A-to-I RNA editing is most commonly observed in coding mRNAs from more primitive organisms (2-5) or repetitive sequences in mammals (6-8), whereas m6A is widespread in the transcriptomes of many animal species (9-17). In humans, tens of thousands of m6A sites have been identified in cell lines, such as HeLa cells (13,14); typically, these sites are enriched around stop codons and have the consensus sequence context GRAC (R = A or G; A = methylated A) (14). Proteins interacting with m6A include the m6A writers (METTL3 and METTL14), readers (YTH domain proteins) and erasers (18). Functional studies have shown that m6A can affect mRNA stability (10,13) or promote translation of host genes (10,14) according to the different readers that bind to m6A sites. Interestingly, in human HeLa cells, m6A sites bound by the reader protein YTHDF1 increase the translation efficiency of host genes (14), whereas m6A sites bound by another reader (IGF2BP) can either facilitate translation or stabilize host mRNAs (10). A handful of studies have discussed the potential relationship between the m6A modification and cancer (10,19-21), but no study has systematically investigated how m6A can affect the translation of global oncogenes (rather than particular oncogenes). Furthermore, the overall relationship (or overlap) between m6A genes and oncogenes is unreported. We believe that this basic information is important for determining whether m6A genes and oncogenes are mutually favored or avoided. Importantly, even researchers who mention the enhanced translation efficiency of oncogenes caused by m6A have not given a “biological” reason why translation of the target genes should be elevated. Specifically, why should m6A facilitate the translation of oncogenes if the oncogenes are already optimized for a high translation efficiency? With the development of next-generation sequencing (NGS) techniques, NGS big data and many bioinformatics tools have been commonly applied to cancer studies (22,23). In this study, by utilizing the YTHDF1 target genes and the normal or si-METTL3 NGS data from HeLa cells generated by a previous work (14), we demonstrate that the m6A genes in HeLa cells originally are unfavorable for translation due to their significantly longer mRNA lengths and stronger codon usage bias. However, the translation of these target genes is compensated by the m6A modification, because the translation efficiency of the m6A genes is significantly higher in normal cells (with m6A) than in si-METTL3 cells (no m6A). Furthermore, we found enrichment of m6A genes in human oncogenes, and these oncogenes were even less suitable for translation. Our results suggest that the compensation of translation by m6A may originally have been designed for those oncogenes to help cancer cell growth. Alternatively, the m6A modification at least facilitates the translation of target genes that originally are unfavorable for translation. This strategy may be used by cancer cells to increase the protein quantity of a particular set of genes and eventually achieve rapid cell growth. This study deepened our understanding of the role played by m6A modification in cancer cells and revealed why m6A genes and some oncogenes need methylation to enhance their translation. We also provided novel insights into a potential method to suppress oncogenes in cancer cells.
Methods
Data collection
We collected the m6A peaks bound by the reader protein YTHDF1 reported in a previous study (14). The list of oncogenes was downloaded from the latest version of the Cancer Gene Census website (CGC, https://cancer.sanger.ac.uk/census/). NGS mRNA-Seq and Ribo-Seq data from normal or si-METTL3 HeLa cells were obtained from the same study (14). Adenosines within the GRAC (R = A or G; A = methylated A) motif in m6A peaks were defined as m6A sites in HeLa cells.
Annotation of m6A sites
We annotated the m6A sites using the hg19 human genome downloaded from the UCSC Genome Browser (genome.ucsc.edu). If an m6A site hit multiple isoforms of the same gene, then the transcript with the longest CDS (canonical transcript) was retained. The canonical transcript of each gene was defined by the SnpEff software (24). An m6A modification that did not hit any genes was annotated as intergenic.
Processing of the next-generation sequencing data
We aligned the NGS reads (mRNA-Seq and Ribo-Seq from the normal and si-METTL3 cells) to the hg19 reference genome using STAR (25). The uniquely mapped reads were kept for downstream analysis. The read counts of each gene in each sample were calculated by htseq-count (26). In the gene expression analysis, the canonical transcript of each gene was chosen, and all reads that overlapped with exon regions were counted.
Calculating differences in translation efficiency
We counted the reads within CDS regions of the canonical transcripts of each gene. The translation efficiency (TE) was defined as the ratio of normalized Ribo-Seq and mRNA-Seq read counts. Here, using the mRNA-Seq and Ribo-Seq counts from the normal (Control) and si-METTL3 (Treated) conditions, we employed xtail (27) to detect differences in the translation efficiency between the Control and Treated samples. The TE of the Control and Treated conditions and the log2TE fold change (FC) were given by the software. Genes with a log2TE FC <0 represent genes with a downregulated TE following si-METTL3 treatment.
Gene ontology (GO) enrichment
The GO analysis was performed using DAVID (28). Highly expressed genes (raw read count >100) in normal HeLa cells were used as background genes.
Conservation analysis
The conservation level of genomic positions is measured by the phyloP score (downloaded from the UCSC Genome Browser, genome.ucsc.edu). Briefly, sites with higher conservation levels have higher phyloP scores. For the comparison of the conservation levels of m6A+ and m6A− sites in coding regions, sites in different codon positions were compared separately.
Codon usage bias
The protocol used to calculate codon usage bias was described in an earlier study (29). The codon bias of a gene is calculated by the deviation (chi-square) of the A/T content of synonymous codons from that of the intronic regions. Higher deviation indicates a stronger codon bias for a gene. The codon bias of a particular codon is the correlation coefficient between the codon frequency within a synonymous codon family and the deviation (Chi-square value) of each gene (29). A higher correlation coefficient suggests stronger bias for a codon.
Statistical analysis
All statistical analyses were conducted in the R environment (http://www.R-project.org/).
Results
m6A methylome in human HeLa cells
We retrieved the m6A peaks (YTHDF1 target) identified in HeLa cells from a previous study (14) and extracted all adenosine sites within the GRAC (R = A or G; A = methylated A) motif according to the instructions in the literature. Adenosines located in the GRAC context in m6A peaks were regarded as m6A sites (Figure 1A). In total, we obtained 18,276 unique m6A sites. We annotated these m6A sites according to the reference genome hg19, and the canonical transcript of each gene was chosen if a site was located in multiple isoforms (Methods). The majority of m6A sites were located in CDS regions, and the 3'UTRs (untranslated regions) also contained a large fraction of m6A sites (Figure 1B). Apart from a few m6A sites in intergenic regions, most of the sites were assigned to 6,025 unique human genes. The m6A genes were significantly enriched in transcription factors based on the GO enrichment analysis (Figure 1C, only GO terms with FDR values <0.05 were listed), which agreed well with known concepts.
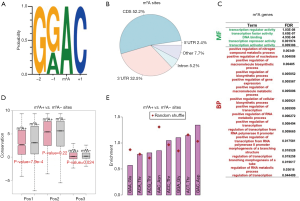
The m6A modification was previously reported to increase the translation efficiency of host genes (14,17). Since m6A events (most of which are located around stop codons) have been reported to help recruit translation initiation factors and facilitate translation, the exact position of an m6A site on a mRNA may not be important as long as the methylation event takes place on this mRNA (in the CDS, UTRs or around stop codons). To investigate whether the particular m6A position was important, we sought potential differences between m6A sites (m6A+) and comparable non-m6A sites (m6A−). The m6A− sites were defined as unmethylated adenosines within GRAC motifs in m6A genes (in HeLa cells). We found that the m6A+ sites in coding regions largely subjected to natural selection were not more conserved than the m6A− sites at the genome level (Figure 1D and Methods). This result agreed with an earlier study, which reported that generally human m6A sites were non-conserved (30). The particular m6A positions may not be important; otherwise, they would be preserved by natural selection and exhibit high conservation levels. Furthermore, we compared codons containing m6A+ and m6A− sites in CDSs. Due to the constraint of the GRAC sequence context, the m6A sites were only found in a small set of codons, and the m6A+ sites did not show any striking enrichment compared to the m6A− sites (Figure 1E). Again, we could not deduce any putative function of the m6A positions from this result, suggesting that the m6A modifications might not exert their function at the “intra-gene” level. Instead, as many previous studies have revealed (14,17), the major function of m6A is to increase the translation efficiency of host genes.
Crosstalk between m6A genes and oncogenes
m6A methylation events are known to promote host gene translation (14,17), and this mechanism does not rely much on the exact position of the methylation sites. Next, we investigated whether human cancer cells could enhance the translation of oncogenes via m6A modification.
To address this question, first we searched for known human oncogenes from the Cancer Gene Census (CGC, https://cancer.sanger.ac.uk/census/). We downloaded 719 human oncogenes from the latest version of the CGC website. A total of 335 of these 719 oncogenes were methylated in HeLa cells (Figure 2A). Furthermore, considering the mRNA expression levels, we extracted genes with a raw read count >100 in HeLa cells (see Methods for detail). A total of 4,968 of the 6,025 m6A genes and 478 of the 719 oncogenes were highly expressed in HeLa cells. The 4,968 m6A genes and 478 oncogenes included 283 overlapping genes (Figure 2B). Thus, more than half of the highly expressed oncogenes were methylated. Then, we examined the gene ontology of the highly expressed oncogenes. We found that these oncogenes were enriched in the transcriptional regulation and metabolism categories (Figure 2C), which agreed with the known features of m6A genes. Next, we calculated the number and density of m6A sites in oncogenes and other genes. To exclude the potential bias caused by mRNA length (as longer genes tend to bear more m6A sites by chance), we ranked all genes into five groups with decreasing mRNA length. Within each bin, we compared the number of m6A sites (Figure 2D, top) and the density of m6A sites (Figure 2D, bottom) in oncogenes versus other genes. The m6A density is defined as m6A sites per adenosine, which canceled the bias introduced by gene length. Strikingly, our results show that both the m6A number and density is generally higher in oncogenes than other genes (Figure 2D). This pattern indicates a potential functional role of the m6A modification in oncogenes.
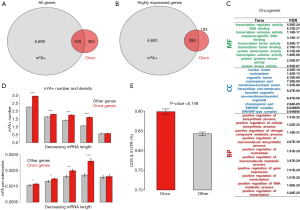
We began to search for differences between the oncogenes and non-oncogenes (termed other genes) or between the m6A genes and non-m6A genes. First, we compared the m6A site distribution of the oncogenes and the remaining genes among the methylated gene set. We calculated the proportion of m6A sites that were located in CDSs and 3'UTRs. Intriguingly, the oncogenes showed a remarkably higher fraction of m6A sites in the CDSs and 3'UTRs than the other genes (Figure 2E). Since YTHDF1 binding of m6A sites facilitate translation via recruitment of initiation factors and circularization of host mRNAs (14), the m6A modifications on CDSs and 3'UTRs are likely to assist with the recruitment and circularization processes (than those modifications in regions such as the 5'UTR).
m6A genes, including oncogenes, are unfavorable for translation
Circularization of mRNA is important for translation. The possibility that the mRNA length may be an important factor that influences circularization is intuitive. We speculated that the longer genes had more difficulty with circularization and therefore were less favorable for translation (this assumption is tested in the next section). Among the highly expressed m6A genes in HeLa cells, we globally profiled the relationship between the mRNA length (the genes were divided into bins) and the fraction of m6A sites in the CDSs and 3'UTRs. Interestingly, the fraction of m6A sites in the CDSs and 3'UTRs increased with the mRNA length (Figure 3A, P<2.2e-16). This result suggests that longer m6A genes have a greater need to promote their translation by methylation. Next, we compared the mRNA lengths of the m6A genes versus non-m6A genes (Figure 3B) or oncogenes versus other genes (Figure 3C). The results are as follows: (I) m6A genes are significantly longer than non-m6A genes (Figure 3B), and (II) among these two gene sets, the oncogenes are significantly longer than the other genes (Figure 3C). If longer genes are indeed unfavorable for translation, then the m6A genes (and especially the oncogenes among them) will suffer from a disadvantage in translation.
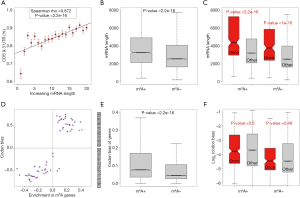
Another factor that can influence the translation rate is (synonymous) codon usage bias. During translation elongation, the rate-limiting step of the decoding process is waiting for the corresponding tRNA of each codon. A relationship should exist between codon usage and the translation efficiency. We followed the method used an early study (29) to calculate the codon bias of each human gene and each codon (see Methods for details). For m6A genes versus non-m6A genes in HeLa cells, we found that the codons enriched in the m6A genes had a stronger bias (Figure 3D). At the gene level, the m6A genes had a stronger bias than the non-m6A genes (Figure 3E), whereas the oncogenes showed almost no difference in codon bias compared to that of the other genes (Figure 3F). If a stronger codon bias is unfavorable for translation (tested in the following section), then the m6A genes should find a solution to neutralize this disadvantage.
m6A methylation facilitates the translation of host genes, including oncogenes
We have proposed that a longer mRNA length and stronger codon bias may be unfavorable for translation and that m6A genes may suffer from these disadvantages. Here, using mRNA-Seq and Ribo-Seq NGS data from HeLa cells generated by a previous study (14), we examined the correlation between the translation efficiency and CDS length (Figure 4A) or codon bias (Figure 4B). Both variables show a negative correlation with the translation efficiency (P<2.2e-16). These correlations verified our assumption that both long mRNA (CDS) lengths and strong codon bias were unfavorable for translation. Moreover, the length and codon bias did not show any correlations (Figure 4C), proving that these two factors might contribute independently to the lower translation efficiency.
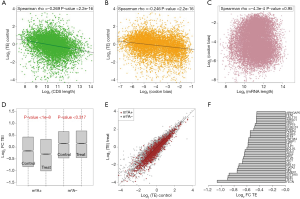
A question arises that since m6A genes (especially the oncogenes) are less suitable for translation, can they compensate for these disadvantages by m6A modification? We fully utilized data from normal (Control) and si-METTL3 (Treated) HeLa cells. Knock down (si-) of the m6A writer gene METTL3 largely reduced the transcriptome-wide m6A level (14). We compared the translation efficiency of m6A and non-m6A genes in normal or si-METTL3 HeLa cells. As expected, the m6A genes but not the non-m6A genes showed a reduced translation efficiency in the si-METTL3 condition (Figure 4D,E). Notably, the translation efficiency of the m6A genes was remarkably lower than that of the non-m6A genes when the m6A writer was removed, but the translation efficiencies of the m6A genes were still lower, even with help from m6A (in the normal condition) (Figure 4D). Take together with our previous results, we propose that the m6A genes are unfavorable for translation due to their longer lengths and stronger codon bias and that they indeed have low translation efficiencies. With the help of the m6A modification, translation of the target genes is elevated.
Since we showed that m6A genes were enriched in oncogenes (Figure 2D), we searched for oncogenes that benefited from m6A modification. We listed the oncogenes with the most decreased translation efficiencies in the si-METTL3 versus normal condition (Figure 4F). This set of oncogenes increased their translation efficiencies through m6A methylation and might play important roles in cancer cell oncogenesis.
Discussion
m6A methylation participates in many biological processes, of which one of the most well-studied functions is facilitating the translation of host genes via the reader protein YTHDF1 (14). Although several studies have mentioned the role of m6A in oncogenesis (10,19,20), they are either case studies of particular genes or do not systematically investigate the translation of target genes. In this work, we utilized the YTHDF1 target genes and normal or si-METTL3 NGS data (mRNA-Seq and Ribo-Seq) in HeLa cells generated by a previous work (14) and elucidated the potential function of the m6A modification in cancer cells.
We found that the m6A genes were enriched in oncogenes when compared to non-m6A genes. We observed remarkably longer mRNA lengths for m6A genes, especially the oncogenes among them. We also observed stronger codon usage bias for m6A genes than for non-m6A genes. Lines of evidence revealed that a longer mRNA length and stronger codon bias were unfavorable for translation, because these two factors were significantly negatively correlated with the translation efficiencies of the genes. This finding provides a simple explanation for why m6A genes (or the oncogenes among them) need the m6A modification to enhance their translation. Indeed, the translation efficiency of m6A genes is significantly elevated in normal HeLa cells (Control) compared to that in si-METTL3 HeLa cells (Treated) where methylation is removed, whereas the translation of non-m6A genes is almost unchanged in the control versus treated cells. In other words, the unfavorable features for translation of m6A genes (or the oncogenes among them) are compensated by the m6A modification (Figure 5). Recruitment of initiation factors does not directly resolve the codon bias problem, but the increased translation initiation rate will definitely enhance the global translation efficiency of host genes. We should note that the impact of codon usage bias on the mRNA translation efficiency is still debatable (31). However, we do not wish to contradict any previous reports. Conservatively, we declare that the translation efficiency of a gene is negatively correlated with its codon bias, at least for the HeLa cell data used in this study.
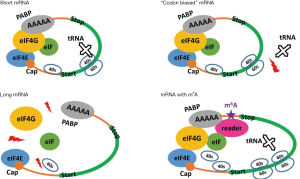
Notably, compensation by m6A modification may originally have been designed for oncogenes, since we have observed enrichment of m6A genes in oncogenes. If methylated oncogenes obtain higher translation efficiencies and eventually facilitate cancer cell proliferation, this strategy or mechanism may explain how cancer cells/tissues achieve rapid cell growth. However, direct evidence for this assumption is still lacking. Hopefully, detailed experimental validation will be carried out in the future.
The main contribution of this study is to unveil the enrichment of m6A genes in oncogenes and to clarify why the m6A target is needed to enhance their translation efficiencies by m6A methylation (Figure 5). Understanding this relationship is important and should be interesting for the fields of RNA modification, translational regulation and cancer studies.
Conclusions
Our results demonstrate that HeLa cells compensate genes unfavorable for translation by m6A modification and enhance their translation efficiencies (Figure 5). These m6A target genes are enriched in oncogenes, and the enhancement of translation of these genes may be related to cancer cell oncogenesis.
Acknowledgments
Funding: This research was financially supported by
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.03.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cantara WA, Crain PF, Rozenski J, et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res 2011;39:D195-201. [Crossref] [PubMed]
- Buchumenski I, Bartok O, Ashwal-Fluss R, et al. Dynamic hyper-editing underlies temperature adaptation in Drosophila. PLoS Genet 2017;13:e1006931. [Crossref] [PubMed]
- Liscovitch-Brauer N, Alon S, Porath HT, et al. Trade-off between Transcriptome Plasticity and Genome Evolution in Cephalopods. Cell 2017;169:191-202.e11. [Crossref] [PubMed]
- Yu Y, Zhou H, Kong Y, et al. The Landscape of A-to-I RNA Editome Is Shaped by Both Positive and Purifying Selection. PLoS Genet 2016;12:e1006191. [Crossref] [PubMed]
- Zhang R, Deng P, Jacobson D, et al. Evolutionary analysis reveals regulatory and functional landscape of coding and non-coding RNA editing. PLoS Genet 2017;13:e1006563. [Crossref] [PubMed]
- Porath HT, Knisbacher BA, Eisenberg E, et al. Massive A-to-I RNA editing is common across the Metazoa and correlates with dsRNA abundance. Genome Biol 2017;18:185. [Crossref] [PubMed]
- Ramaswami G, Lin W, Piskol R, et al. Accurate identification of human Alu and non-Alu RNA editing sites. Nat Methods 2012;9:579-81. [Crossref] [PubMed]
- Ramaswami G, Zhang R, Piskol R, et al. Identifying RNA editing sites using RNA sequencing data alone. Nat Methods 2013;10:128-32. [Crossref] [PubMed]
- Haussmann IU, Bodi Z, Sanchez-Moran E, et al. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 2016;540:301-4. [Crossref] [PubMed]
- Huang H, Weng H, Sun W, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 2018;20:285-95. [Crossref] [PubMed]
- Kan L, Grozhik AV, Vedanayagam J, et al. The m(6)A pathway facilitates sex determination in Drosophila. Nat Commun 2017;8:15737. [Crossref] [PubMed]
- Lence T, Akhtar J, Bayer M, et al. m(6)A modulates neuronal functions and sex determination in Drosophila. Nature 2016;540:242-7. [Crossref] [PubMed]
- Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014;505:117-20. [Crossref] [PubMed]
- Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015;161:1388-99. [Crossref] [PubMed]
- Xu K, Yang Y, Feng GH, et al. Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res 2017;27:1100-14. [Crossref] [PubMed]
- Zheng Y, Nie P, Peng D, et al. m6AVar: a database of functional variants involved in m6A modification. Nucleic Acids Res 2018;46:D139-45. [Crossref] [PubMed]
- Zhou J, Wan J, Gao X, et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015;526:591-4. [Crossref] [PubMed]
- Zhao BS, Nachtergaele S, Roundtree IA, et al. Our views of dynamic N(6)-methyladenosine RNA methylation. RNA 2018;24:268-72. [Crossref] [PubMed]
- Liu J, Eckert MA, Harada BT, et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol 2018;20:1074-83. [Crossref] [PubMed]
- Li Z, Weng H, Su R, et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)- Methyladenosine RNA Demethylase. Cancer Cell 2017;31:127-41. [Crossref] [PubMed]
- Lin S, Choe J, Du P, et al. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell 2016;62:335-45. [Crossref] [PubMed]
- Cao Y, Cong H, Wang XD, et al. Circular RNAs in cancer: old tree with new flowers. Translational Cancer Research 2017;6:843-51. [Crossref]
- Lee CY, Chiu YC, Wang LB, et al. Common applications of next-generation sequencing technologies in genomic research. Translational Cancer Research 2013;2:33-45.
- Cingolani P, Platts A. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80-92. [Crossref] [PubMed]
- Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15-21. [Crossref] [PubMed]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166-9. [Crossref] [PubMed]
- Xiao Z, Zou Q, Liu Y, et al. Genome-wide assessment of differential translations with ribosome profiling data. Nat Commun 2016;7:11194. [Crossref] [PubMed]
- Jiao X, Sherman BT. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics 2012;28:1805-6. [Crossref] [PubMed]
- Akashi H. Inferring weak selection from patterns of polymorphism and divergence at "silent" sites in Drosophila DNA. Genetics 1995;139:1067-76. [PubMed]
- Liu Z, Zhang J. Most m6A RNA modifications in protein-coding regions are evolutionarily unconserved and likely nonfunctional. Mol Biol Evol 2017; [Epub ahead of print]. [PubMed]
- Yang JR, Chen X, Zhang J. Codon-by-codon modulation of translational speed and accuracy via mRNA folding. PLoS Biol 2014;12:e1001910. [Crossref] [PubMed]


