
BRCA1/2 mutation spectrum in Chinese early-onset breast cancer
Introduction
Breast cancer is the most commonly diagnosed cancer among women and is the second leading cause of cancer death for women in US (1). Early-onset breast cancer, though only accounting for 7% of all breast cancers, is the most common cancer among young females (2), and has been described to be more biologically aggressive than in older women, which has been associated with a worse prognosis (3). Nowadays, with the development of new techniques, an increasing number of susceptibility gene mutations related to early-onset breast cancer has been detected to improve diagnosis and therapy of early-onset breast cancer and predict outcome. Among all these detected mutations, BRCA1/2 are still figured out to play an important role in early-onset breast cancer (4-7).
Among the multitude of markers involved into the breast cancer tumorigenesis as EGFR, RANK (8), BRCA1 and BRCA2 are the first two genes found directly related to hereditary breast cancer. BRCA1 is located on 17q21.31, and the exon count is 24. BRCA2 is located on 13q13.1, and the exon count is 27. Both BRCA1 and BRCA2 are considered tumor suppressor gene which involved in maintenance of genome stability, specifically the homologous recombination pathway for double-strand DNA repair. Inherited mutations in BRCA1 and BRCA2, confer increased lifetime risk of developing breast cancer (9). Due to the length of BRCA1/2, and the randomness of mutation sites, using the traditional sanger sequencing, qPCR, or MLPA to detect the whole gene is not ideal. In contrast, the high throughput next-generation sequencing (NGS) could be more efficient, that can detect all the exons and their adjacent regions of BRCA1/2 at a time.
As is mentioned in several published research (10-13), compared with other countries, Chinese women carry different BRCA mutations rate and types. In this study, we tried to figure out a detailed BRCA1/2 germline and somatic mutation spectrum in young Chinese breast cancer patients.
Methods
Cases and samples
A total of 54 female patients diagnosed with breast cancer were enrolled in this study, of which 27 patients (mean age 32 years, range, 23–40 years) diagnosed at the age younger than 40 and the rest 27 (mean age 52 years, range, 41–68 years) diagnosed at the age older than 40 in West China Hospital from January 2010 to December 2016 consecutively, belonging to study group and control group, respectively. DNA of 54 FFPE samples of cancer tissue were collected to test the somatic BRCA1/2 mutations, while DNA of 31 blood samples and 23 FFPE samples of normal tissue were used to exclude the germline BRCA1/2 mutations by two NGS platforms PGM and Miseq. Clinicopathological characteristics were reviewed including age, estrogen-receptor (ER) status, progesterone-receptor (PR) status, human epidermal growth factor-2 (HER-2) status, Ki-67, molecular phenotypes, TNM staging, etc. Patients’ clinical information is showed in Table 1. Approval for the study was granted by the Ethics Committee of West China Hospital (number: 2013-191).
Table 1
| Characteristics | Study group (n=27) | Control group (n=27) | P value |
|---|---|---|---|
| Mean age (years) | 32 | 52 | <0.001 |
| Estrogen-receptor (ER) status | 0.704 | ||
| Positive | 24 | 22 | |
| Negative | 3 | 5 | |
| Progesterone-receptor (PR) status | 0.750 | ||
| Positive | 21 | 20 | |
| Negative | 6 | 7 | |
| Human epidermal growth factor-2 (HER-2) | 0.669 | ||
| Positive | 23 | 25 | |
| Negative | 4 | 2 | |
| Molecular phenotypes | 0.525 | ||
| Luminal A | 1 | 1 | |
| Luminal B (HER2+) | 22 | 22 | |
| Luminal B (HER2−) | 2 | 0 | |
| HER2+ | 2 | 3 | |
| Triple negative | 0 | 1 | |
| TNM stage | |||
| T stage (1 missing) | 0.551 | ||
| Tis | 1 | 0 | |
| T1 | 8 | 8 | |
| T2 | 15 | 17 | |
| T3 | 3 | 1 | |
| T4 | 0 | 0 | |
| N stage (1 missing) | 0.273 | ||
| N0 | 14 | 14 | |
| N1 | 9 | 7 | |
| N2 | 0 | 3 | |
| N3 | 4 | 2 | |
Next generation sequencing on PGM platform and Miseq platform
DNA was extracted using QIAamp DNA FFPE Tissue Kit. For library construction, 30 ng of gDNA (measured using the Qubit fluorometer in combination with the Qubit dsDNA HS assay kit) was amplified using BRCAimPLUS DNA panel (SINGLERA Genomics Inc.) and the Ion Ampliseq™ HiFi Master Mix (Ion Ampliseq™ Library kit 2.0). The amplicons were then digested, barcoded and amplified using the Ion Ampliseq™ Library kit 2.0, Ion Xpress™ barcode adapters kit (Life technologies), and Ion-to-Miseq primers according to the manufacturer’s instructions. The library size was checked using the Agilent High Sensitivity DNA Kit by the Bioanalyzer 2100 instrument (Agilent Technologies), and library concentration was evaluated using the Qubit fluorometer and the Qubit dsDNA HS assay kit (Life technologies).
For PGM platform, 50 pM of each library was multiplexed and clonally amplified on the Chef instrument with the Ion PGM™ Hi-Q™ Chef Solutions Cartridge, Ion PGM™ Hi-Q™ Chef Reagents Cartridge, Ion PGM™ Hi-Q™ Chef Supplies and Ion 318™ Chip v2 breast cancer (Life technologies) according to the manufacturer’s instructions. Finally, the Ion 318™ chips loaded with enriched template ISP were sequenced on a PGM™ sequencer with the Ion PGM™ sequencing 200 kit v2 according to the manufacturer’s instructions.
For Miseq platform, all purified libraries were quantified by real-time PCR using the SYBR Fast Illumina Library Quantification Kit (Kapa Biosystems) and pooled to give equal genome coverage from each library. Each multiplexed library pool was sequenced an Illumina MiSeq for 151 cycles from each end read according to the manufacturer’s instructions.
We then analyzed the sequencing data from both Miseq and PGM platforms using the BRCAimPLUS DNA pipeline-a customized bioinformatic analysis workflow for cancer panel. As other bioinformatic pipelines, it involves processing a series of data transformation steps: alignment, variant calling, annotation, filtering and reporting (14-17).
Variants confirmation
DNA was extracted using QIAamp DNA FFPE Tissue Kit. PCR reactions were run in final volumes of 25 µL containing 200 ng DNA, 0.25 mM dNTPs, 10pmol of each primer and 1.25 unit of Taq polymerase [TIANGEN BIOTECH (BEIJING) CO., LTD.]. PCR was performed in an T100 thermal cycler (Bio-Rad, Hercules, CA, USA) with initial denaturation at 95 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s. The purified PCR products were sequenced by Sanger’s sequencing according to manufacturer’s instruction.
Variants evaluation
Variants both detected by two platforms were then to be evaluated. According to the classification system of International Agency for Research on Cancer (IARC), American College of Medical Genetics and Genomics (ACMG), and Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA), the mutations of BRCA gene were divided into five categories: pathogenic (Class 5, the rate of pathogenicity is higher than 0.99), likely pathogenic (Class 4, the rate of pathogenicity is between 0.95 and 0.99), uncertain significance (Class 3, the rate of pathogenicity is between 0.05 and 0.949), likely benign (Class 2, the rate of pathogenicity is between 0.001 and 0.049), benign (Class 1, the rate of pathogenicity is lower than 0.001).
Also, the BRCA gene variants identified were checked for pathogenicity in 4 databases: Breast Cancer Information Core (BIC) (18), Leiden Open Variation Database (LOVD) (19), the Catalogue of Somatic Mutations in Cancer database (COSMIC) (20) and ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/).
Statistical analysis
All the statistical analyses were performed using the statistical program for social sciences (SPSS) software package version 19.0 (Chicago, IL, USA). Two independent sample t tests were applied in comparison between groups. Enumeration data was expressed as cases or percentage. Chi-square test was used in comparison between groups, with P<0.05 represented for the difference was statistically significant.
Results
NGS test performance
In the cohort of 54 patients, we obtained an average of 4.1 million reads per sample, with a mean coverage of 90% at a mean X coverage of 2031X on the PGM platform and 5.1 million reads per sample with a mean coverage of 92% at a mean X coverage of 2543X on the Miseq platform. In the early-onset breast cancer patients, 2 had no mutations in BRCA1/2 genes. In the rest 25 patients, a total of 12 mutations of BRCA1 were detected by both PGM and Miseq platform in 19 patients (Table 2). Eleven mutations of BRCA2 gene were detected in 22 patients (Table 3). In control group, two patients had no BRCA mutations, while 7 BRCA1 mutations were detected in 22 patients (Table 2), and 9 BRCA2 mutations were detected in 22 patients (Table 3).
Table 2
| Groups | Exon | Intron | Type | Consequence | cDNA_change | Pro_change | Germline mutation (n) | Somatic mutation (n) |
|---|---|---|---|---|---|---|---|---|
| Study group | ||||||||
| BRCA1 | − | 4/23 | SNP | Splice_acceptor_variant | c.213-1G>A | − | 0 | 1 |
| BRCA1 | 10/24 | − | SNP | Missense_variant | c.988G>A | p.Asp330Asn | 1 | 0 |
| BRCA1 | 10/24 | − | SNP | Missense_variant | c.1036C>T | p.Pro346Ser | 1 | 0 |
| BRCA1 | 10/24 | − | Indel | Frameshift_variant | c.1299dupC | p.Ser434GlnfsTer2 | 1 | 0 |
| BRCA1 | 10/24 | − | SNP | Stop_gained | c.2059C>T | p.Gln687Ter | 1 | 0 |
| BRCA1 | 10/24 | − | SNP | Missense_variant | c.2566T>C | p.Tyr856His | 4 | 0 |
| BRCA1 | 10/24 | − | SNP | Missense_variant | c.2612C>T | p.Pro871Leu | 14 | 1 |
| BRCA1 | 10/24 | − | SNP | Missense_variant | c.2623C>T | p.Pro875Ser | 1 | 0 |
| BRCA1 | 10/24 | − | SNP | Missense_variant | c.3113A>G | p.Glu1038Gly | 14 | 1 |
| BRCA1 | 10/24 | − | SNP | Missense_variant | c.3548A>G | p.Lys1183Arg | 14 | 1 |
| BRCA1 | 15/24 | − | SNP | Splice_region_variant&synonymous_variant | c.4674A>G | c.4674A>G (p.Leu1558=) | 1 | 0 |
| BRCA1 | 16/24 | − | SNP | Missense_variant | c.4837A>G | p.Ser1613Gly | 15 | 0 |
| Control group | ||||||||
| BRCA1 | 7/24 | − | SNP | Missense_variant | c.446A>C | p.Glu149Ala | 1 | 0 |
| BRCA1 | 10/24 | − | Indel | Frameshift_variant | c.2398_2401delAAAT | p.Lys800ValfsTer2 | 0 | 1 |
| BRCA1 | 10/24 | − | SNP | Missense_variant | c.2566T>C | 10 | 1 | 0 |
| BRCA1 | 10/24 | − | SNP | Missense_variant | c.2612C>T | p.Pro871Leu | 16 | 2 |
| BRCA1 | 10/24 | − | SNP | Missense_variant | c.3113A>G | p.Glu1038Gly | 17 | 4 |
| BRCA1 | 10/24 | − | SNP | Missense_variant | c.3548A>G | p.Lys1183Arg | 17 | 2 |
| BRCA1 | 16/24 | − | SNP | Missense_variant | c.4837A>G | p.Ser1613Gly | 17 | 3 |
Table 3
| Groups | Exon | Type | Consequence | cDNA_change | Pro_change | Germline mutation (n) | Somatic mutation (n) |
|---|---|---|---|---|---|---|---|
| Study group | |||||||
| BRCA2 | 27/27 | SNP | Missense_variant | c.10234A>G | p.Ile3412Val | 0 | 1 |
| BRCA2 | 25/27 | Indel | Frameshift_variant | c.9401delG | p.Gly3134AlafsTer29 | 1 | 0 |
| BRCA2 | 18/27 | SNP | Missense_variant | c.8187G>T | p.Lys2729Asn | 1 | 0 |
| BRCA2 | 11/27 | SNP | Missense_variant | c.5852G>A | p.Ser1951Asn | 1 | 0 |
| BRCA2 | 11/27 | SNP | Missense_variant | c.5785A>G | p.Ile1929Val | 1 | 0 |
| BRCA2 | 11/27 | SNP | Missense_variant | c.2971A>G | p.Asn991Asp | 6 | 0 |
| BRCA2 | 10/27 | SNP | Missense_variant | c.1462A>G | p.Ile488Val | 1 | 0 |
| BRCA2 | 10/27 | SNP | Stop_gained | c.1399A>T | p.Lys467Ter | 1 | 0 |
| BRCA2 | 10/27 | SNP | Missense_variant | c.1114A>C | p.Asn372His | 13 | 1 |
| BRCA2 | 10/27 | SNP | Missense_variant | c.865A>C | p.Asn289His | 6 | 0 |
| BRCA2 | 5/27 | SNP | Missense_variant | c.461A>G | p.Gln154Arg | 1 | 0 |
| Control group | |||||||
| BRCA2 | 27/27 | SNP | Missense_variant | c.10234A>G | p.Ile3412Val | 3 | 0 |
| BRCA2 | 27/27 | SNP | Missense_variant | c.10150C>G | p.Arg3384Gly | 1 | 0 |
| BRCA2 | 25/27 | SNP | Stop_gained | c.9294C>G | p.Tyr3098Ter | 1 | 0 |
| BRCA2 | 14/27 | Indel | Frameshift_variant | c.7414_7415delAA | p.Lys2472ValfsTer2 | 0 | 1 |
| BRCA2 | 11/27 | SNP | Missense_variant | c.5785A>G | p.Ile1929Val | 1 | 0 |
| BRCA2 | 11/27 | SNP | Missense_variant | c.3445A>G | p.Met1149Val | 1 | 0 |
| BRCA2 | 11/27 | SNP | Missense_variant | c.2971A>G | p.Asn991Asp | 7 | 2 |
| BRCA2 | 10/27 | SNP | Missense_variant | c.1114A>C | p.Asn372His | 11 | 6 |
| BRCA2 | 10/27 | SNP | Missense_variant | c.865A>C | p.Asn289His | 7 | 2 |
Germline BRCA1/2 mutations detected in young breast cancer patients
In study group, the 11 BRCA1 germline mutations detected were classified as following: 2 5-pathogenic, 4 3-uncertain, 5 1-benign, in accordance with the data found in ClinVar (Tables S1,S2). c.2623C>T was identified as novel germline mutation site found in a patient diagnosed as breast cancer at the age of 27 (Figure S1A). Among all, 4 mutations were found to be pending, while the rest were not found in BIC. According to the Leiden Open Variation Database, 2 mutations were definitely indicated to affect function. In COSMIC database, 4 mutations were found to be neutral.
As for BRCA2, 10 germline mutations were detected involving 2 5-pathogenic, 3 3-uncertain, 5 1-benign in accordance with the data found in ClinVar (Tables S1,S2). c.5852G>A identified as a novel site found in a patient diagnosed as breast cancer at the age of 28 (Figure S1B). Among all, 1 mutation c.10234A>G was found to be class 1 in BIC database, 2 mutations were found to be pending, while the rest were not found in BIC. According to the Leiden Open Variation Database, 1 mutation was definitely indicated to affect function. In COSMIC database, 4 mutations were found to be neutral, and 1 referring to be pathogenic.
In control group, there were only 6 BRCA1 germline in total 27 patients. c.446A>C was a mutation with uncertain significance. c.2566T>C, c.2612C>T, c.3113A>G, c.3548A>G, c.4837A>G were benign mutations. When considered BRCA2, 1 pathogenic mutation c.9294C>G, 2 uncertain mutations c.10150C>G, c.3445A>G were identified as well as 5 benign mutations.
Somatic BRCA1/2 mutations detected in young breast cancer patients
In study group, there were 4 BRCA1 somatic mutations detected in total, including pathogenic mutation c.213-1G>A and three benign mutation c.2612C>T, c.3113A>G, c.3548A>G only detected in one patient respectively. Also, two BRCA2 somatic benign mutations c.10234A>G and c.1114A>C were detected.
In control group, 5 BRCA1 somatic mutations involving pathogenic mutation c .2398_2401delAAAT detected in one patient, and four benign mutations c.2612C>T, c.3548A>G, c.4837A>G, c.3113A>G. Also, 4 BRCA2 somatic mutations were detected, of which c.7414_7415delAA was pathogenic with the rest c.2971A>G, c.865A>C, and c.1114A>C turned out to be benign.
BRCA1/2 mutations of pathogenic and uncertain significance in young breast cancer patients
Mutations defined as pathogenic/likely pathogenic and uncertain were selected for further analysis, as is shown in Table 4. In total, 11 germline (7 3-uncertain, 4 5-pathogenic) and 1 somatic (5-pathogenic) of BRCA1/2 mutations were detected in study group, while 4 germline (3 3-uncertain and 1 5-pathogenic) and 2 somatic (2 5-pathogenic) of BRCA1/2 mutations were detected in control group.
Table 4
| Groups | Variations | Clinical significance | Germline mutations (n) | Somatic mutations (n) |
|---|---|---|---|---|
| Study group | ||||
| BRCA1 | c.988G>A | 3-uncertain | 1 | 0 |
| BRCA1 | c.4674A>G | 3-uncertain | 1 | 0 |
| BRCA1 | c.1036C>T | 3-uncertain | 1 | 0 |
| BRCA1 | c.2623C>T | 3-uncertain | 1 | 0 |
| BRCA1 | c.2059C>T | 5-pathogenic | 1 | 0 |
| BRCA1 | c.1299dupC | 5-pathogenic | 1 | 0 |
| BRCA1 | c.213-1G>A | 5-pathogenic | 0 | 1 |
| BRCA2 | c.1462A>G | 3-uncertain | 1 | 0 |
| BRCA2 | c.461A>G | 3-uncertain | 1 | 0 |
| BRCA2 | c.5852G>A | 3-uncertain | 1 | 0 |
| BRCA2 | c.9401delG | 5-pathogenic | 1 | 0 |
| BRCA2 | c.1399A>T | 5-pathogenic | 1 | 0 |
| Control group | ||||
| BRCA1 | c.446A>C | 3-uncertain | 1 | 0 |
| BRCA1 | c.2398_2401delAAAT | 5-pathogenic | 0 | 1 |
| BRCA2 | c.10150C>G | 3-uncertain | 1 | 0 |
| BRCA2 | c.3445A>G | 3-uncertain | 1 | 0 |
| BRCA2 | c.9294C>G | 5-pathogenic | 1 | 0 |
| BRCA2 | c.7414_7415delAA | 5-pathogenic | 0 | 1 |
In study group, 14.8% (4/27) and 3.7% (1/27) patients had deleterious BRCA1/2 germline and somatic mutations respectively. While in control group, only 3.7% (1/27) and 7.4% (2/27) had deleterious BRCA1/2 germline and somatic mutations, respectively.
The 4 pathogenic germline mutations were c.2059C>T, c.1299dupC, c.9401delG, c.1399A>T found in study group at the age of 40, 28, 36, 40. And 7 uncertain germline mutations existed in study group were c.988G>A, c.4674A>G, c.1036C>T, c.2623C>T, c.1462A>G, c.461A>G, c.5852G>A. When it turns to control group, there was only one pathogenic germline mutation c.9294C>G found in patient at the age of 57 and 3 uncertain germline mutations c.446A>C, c.10150C>G, c.3445A>G.
Discussion
BRCA status is not only important for the identification of familial cancer predisposition but also to therapeutic choices for breast cancer patients, e.g., the PARP inhibitor therapy (21-23). BRCA gene mutation is closely related to the early-onset breast cancer occurrence. In most national and international guidelines, testing criteria of BRCA includes patients with breast cancer aged less than 35 or 40 years (24). BRCA1/2 mutation carriers diagnosed with breast cancer before age 50 are prone to a worse survival (25). In our study, the mean age of 27 early-age onset breast cancer patients was 32, with the minimum hospitalized age 23. Only 2 had no BRCA1/2 mutation. Twenty patients had BRCA1 mutations and 22 had BRCA2 mutations, with 3 (11.1%) patients had pathogenic mutations involving c.1299dupC, c.2059C>T, c.213-1G>A in BRCA1/2 (7.4%) had pathogenic mutations involving c.9401delG, c.1399A>T in BRCA2, which were not found in control group. The mutation frequency of the deleterious germline mutation in our study is a little bit higher than in other research (24,26,27) may attribute to insufficient number of analyzed cases.
BRCA1 mutation c.2623C>T was identified as a germline mutation for the first time in this study, which was a SNP detected in EXON10 leading to the protein change p.Pro875Ser. The patient who had c.2623C>T mutation as the only BRCA mutation diagnosed as breast cancer at the age of 27. As for BRCA2, c.5852G>A was also identified as a germline mutation for the first time in this study which was a SNP detected in EXON11 leading to the protein change p.Ser1951Asn. The patient who had c.5852G>A mutation as the only BRCA mutation other than benign mutations diagnosed as breast cancer at the age of 28. Since these two new mutation sites were detected in patients at such young age without other suspicious BRCA mutation sites, we have reason to suspect that these two mutation sites may play a role in the pathogenesis of the early-onset breast cancer which need to be further confirmed.
A large number of literatures show strong correlation between BRCA mutation and familial early-onset breast cancer (28-30). In our study, in total, 11 germline (7 3-uncertain, 4 5-pathogenic) and 1 somatic (5-pathogenic) of BRCA1/2 mutations were detected in study group, while 4 germline (3 3-uncertain and 1 5-pathogenic) and 2 somatic (2 5-pathogenic) of BRCA1/2 mutations were detected in control group. In study group, 14.8% (4/27) patients had deleterious BRCA1/2 germline mutations, and 3.7% (1/27) had deleterious BRCA1/2 somatic mutations, while in control group, 3.7% (1/27) had deleterious BRCA1/2 germline mutations, and 7.4% (2/27) had deleterious BRCA1/2 somatic mutations, displaying a trend that early-onset group is more likely to have germline mutations than elderly counterparts. Therefore, there is a strong recommendation for the early-onset breast cancer patients despite of the family history to get BRCA test for potential benefit for their family members as well as benefit for the patient of PARP inhibitor therapy.
One of the limitations of the study is the absence of triple negative samples in the study groups which is among the breast cancer histotypes the more aggressive with the poor prognosis caused by insufficient cases, but we tried to figure out a detailed spectrum of BRCA1/2 germline and somatic mutations of early-onset breast cancer patients in West China Hospital using NGS. Several deleterious and uncertain mutations were observed in this cohort and it is recommended that a more thorough and functional examination of these mutations should be conducted in the future.
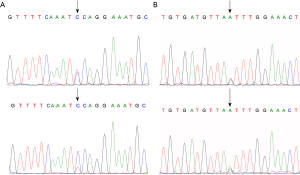
Table S1
| Variations | Clinical significance | BIC database clinically importance/clinical classification | LOVD | COSMIC | ClinVar |
|---|---|---|---|---|---|
| Study group | |||||
| c.213-1G>A | 5-pathogenic | Pending | Affects function | Not found | Pathogenic |
| c.988G>A | 3-uncertain | Not found | Not found | Not found | Uncertain significance |
| c.4674A>G | 3-uncertain | Not found | Effect unknown | Not found | Uncertain significance |
| c.1299dupC | 5-pathogenic | Not found | Not found | Not found | Pathogenic |
| c.2623C>T | 3-uncertain | Not found | Not found | Not found | Not found |
| c.1036C>T | 3-uncertain | Pending | Effect unknown; affects function | Not found | Conflicting interpretations of pathogenicity |
| c.2059C>T | 5-pathogenic | Not found | Affects function | Not found | Pathogenic |
| c.2566T>C | 1-benign | Pending | Does not affect function | Not found | Benign |
| c.2612C>T | 1-benign | Not found | Does not affect function | Neutral | Benign |
| c.3113A>G | 1-benign | Not found | Does not affect function | Neutral | Benign |
| c.3548A>G | 1-benign | Pending | Does not affect function | Neutral | Benign |
| c.4837A>G | 1-benign | Pending | Does not affect function | Neutral | Benign |
| Control group | |||||
| c.446A>C | 3-uncertain | Not found | Not found | Not found | Uncertain significance |
| c.2398_2401delAAAT | 5-pathogenic | Not found | Not found | Not found | Pathogenic |
| c.2566T>C | 1-benign | Pending | Does not affect function | Not found | Benign |
| c.2612C>T | 1-benign | Not found | Does not affect function | Neutral | Benign |
| c.3113A>G | 1-benign | Not found | Does not affect function | Neutral | Benign |
| c.3548A>G | 1-benign | Pending | Does not affect function | Neutral | Benign |
| c.4837A>G | 1-benign | Pending | Does not affect function | Neutral | Benign |
BIC, Breast Cancer Information Core; LOVD, Leiden Open Variation Database; COSMIC, Catalogue of Somatic Mutations in Cancer database.
Table S2
| Variations | Clinical significance | BIC database clinically importance/clinical classification | LOVD | COSMIC | ClinVar |
|---|---|---|---|---|---|
| Study group | |||||
| c.9401delG | 5-pathogenic | Not found | Not found | Not found | Pathogenic |
| c.8187G>T | 1-benign | Pending | Does not affect function | Pathogenic | Benign |
| c.5852G>A | 3-uncertain | Not found | Not found | Not found | Not found |
| c.1462A>G | 3-uncertain | Not found | Not found | Not found | Conflicting interpretations of pathogenicity |
| c.1399A>T | 5-pathogenic | Not found | Affects function | Not found | Pathogenic |
| c.461A>G | 3-uncertain | Not found | Not found | Not found | Uncertain significance |
| c.10234A>G | 1-benign | Class 1 | Does not affect function | Neutral | Benign |
| c.5785A>G | 1-benign | Pending | Does not affect function; Effect unknown | Not found | Benign |
| c.2971A>G | 1-benign | Not found | Does not affect function | Neutral | Benign |
| c.1114A>C | 1-benign | Not found | Does not affect function | Neutral | Benign |
| c.865A>C | 1-benign | Not found | Does not affect function | Neutral | Benign |
| Control group | |||||
| c.10150C>G | 3-uncertain | Not found | Not found | Not found | Uncertain significance |
| c.9294C>G | 5-pathogenic | Class 5 | Affects function | Not found | Pathogenic |
| c.7414_7415delAA | 5-pathogenic | Not found | Not found | Not found | Pathogenic |
| c.3445A>G | 3-uncertain | Pending | Effect unknown | Not found | Conflicting interpretations of pathogenicity |
| c.10234A>G | 1-benign | Class 1 | Does not affect function | Neutral | Benign |
| c.5785A>G | 1-benign | Pending | Does not affect function; Effect unknown | Not found | Benign |
| c.2971A>G | 1-benign | Not found | Does not affect function | Neutral | Benign |
| c.1114A>C | 1-benign | Not found | Does not affect function | Neutral | Benign |
| c.865A>C | 1-benign | Not found | Does not affect function | Neutral | Benign |
BIC, Breast Cancer Information Core; LOVD, Leiden Open Variation Database; COSMIC, Catalogue of Somatic Mutations in Cancer database.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.03.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Approval for the study was granted by the Ethics Committee of West China Hospital (number: 2013-191). And the study was done with all patients’ informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- DeSantis CE, Ma J, Goding Sauer A, et al. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin 2017;67:439-48. [Crossref] [PubMed]
- Anders CK, Johnson R, Litton J, et al. Breast cancer before age 40 years. Semin Oncol 2009;36:237-49. [Crossref] [PubMed]
- Bonnier P, Romain S, Charpin C, et al. Age as a prognostic factor in breast cancer: relationship to pathologic and biologic features. Int J Cancer 1995;62:138-44. [Crossref] [PubMed]
- Antoniou A, Pharoah PD, Narod S, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 2003;72:1117-30. [Crossref] [PubMed]
- Lee DS, Yoon SY, Looi LM, et al. Comparable frequency of BRCA1, BRCA2 and TP53 germline mutations in a multi-ethnic Asian cohort suggests TP53 screening should be offered together with BRCA1/2 screening to early-onset breast cancer patients. Breast Cancer Res 2012;14:R66. [Crossref] [PubMed]
- Moghadasi S, Eccles DM, Devilee P, et al. Classification and Clinical Management of Variants of Uncertain Significance in High Penetrance Cancer Predisposition Genes. Hum Mutat 2016;37:331-6. [Crossref] [PubMed]
- Torrezan GT, de Almeida FGDSR, Figueiredo MCP, et al. Complex Landscape of Germline Variants in Brazilian Patients With Hereditary and Early Onset Breast Cancer. Front Genet 2018;9:161. [Crossref] [PubMed]
- Mercatali L, La Manna F, Miserocchi G, et al. Tumor-Stroma Crosstalk in Bone Tissue: The Osteoclastogenic Potential of a Breast Cancer Cell Line in a Co-Culture System and the Role of EGFR Inhibition. Int J Mol Sci 2017;18:E1655. [Crossref] [PubMed]
- Mann GJ, Thorne H, Balleine RL, et al. Analysis of cancer risk and BRCA1 and BRCA2 mutation prevalence in the kConFab familial breast cancer resource. Breast Cancer Res 2006;8:R12. [Crossref] [PubMed]
- Cao W, Wang X, Li JC. Hereditary breast cancer in the Han Chinese population. J Epidemiol 2013;23:75-84. [Crossref] [PubMed]
- Lang GT, Shi JX, Hu X, et al. The spectrum of BRCA mutations and characteristics of BRCA-associated breast cancers in China: Screening of 2,991 patients and 1,043 controls by next-generation sequencing. Int J Cancer 2017;141:129-42. [Crossref] [PubMed]
- Li G, Guo X, Tang L, et al. Analysis of BRCA1/2 mutation spectrum and prevalence in unselected Chinese breast cancer patients by next-generation sequencing. J Cancer Res Clin Oncol 2017;143:2011-24. [Crossref] [PubMed]
- Kwong A, Ho JCW, Shin VY, et al. Rapid detection of BRCA1/2 recurrent mutations in Chinese breast and ovarian cancer patients with multiplex SNaPshot genotyping panels. Oncotarget 2017;9:7832-43. [PubMed]
- Li B, Krishnan VG, Mort ME, et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 2009;25:2744-50. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491-8. [Crossref] [PubMed]
- Szabo C, Masiello A, Ryan JF, et al. The breast cancer information core: database design, structure, and scope. Hum Mutat 2000;16:123-31. [Crossref] [PubMed]
- Fokkema IF, Taschner PE, Schaafsma GC, et al. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat 2011;32:557-63. [Crossref] [PubMed]
- Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 2015;43:D805-11. [Crossref] [PubMed]
- Tutt A, Robson M, Garber JE, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 2010;376:235-44. [Crossref] [PubMed]
- Plummer R. Poly(ADP-ribose) polymerase inhibition: a new direction for BRCA and triple-negative breast cancer? Breast Cancer Res 2011;13:218. [Crossref] [PubMed]
- Byrski T, Huzarski T, Dent R, et al. Pathologic complete response to neoadjuvant cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res Treat 2014;147:401-5. [Crossref] [PubMed]
- Fasching PA. Breast cancer in young women: do BRCA1 or BRCA2 mutations matter? Lancet Oncol 2018;19:150-1. [Crossref] [PubMed]
- Schmidt MK, van den Broek AJ, Tollenaar RA, et al. Breast Cancer Survival of BRCA1/BRCA2 Mutation Carriers in a Hospital- Based Cohort of Young Women. J Natl Cancer Inst 2017;109:djw329. [Crossref] [PubMed]
- Son BH, Ahn SH, Kim SW, et al. Prevalence of BRCA1 and BRCA2 mutations in non-familial breast cancer patients with high risks in Korea: the Korean Hereditary Breast Cancer (KOHBRA) Study. Breast Cancer Res Treat 2012;133:1143-52. [Crossref] [PubMed]
- Zugazagoitia J, Perez-Segura P, Manzano A, et al. Limited family structure and triple-negative breast cancer (TNBC) subtype as predictors of BRCA mutations in a genetic counseling cohort of early-onset sporadic breast cancers. Breast Cancer Res Treat 2014;148:415-21. [Crossref] [PubMed]
- Loman N, Johannsson O, Kristoffersson U, et al. Family history of breast and ovarian cancers and BRCA1 and BRCA2 mutations in a population-based series of early-onset breast cancer. J Natl Cancer Inst 2001;93:1215-23. [Crossref] [PubMed]
- Semple J, Metcalfe KA, Lubinski J, et al. Does the age of breast cancer diagnosis in first-degree relatives impact on the risk of breast cancer in BRCA1 and BRCA2 mutation carriers? Breast Cancer Res Treat 2015;154:163-9. [Crossref] [PubMed]
- Azzollini J, Scuvera G, Bruno E, et al. Mutation detection rates associated with specific selection criteria for BRCA1/2 testing in 1854 high-risk families: A monocentric Italian study. Eur J Intern Med 2016;32:65-71. [Crossref] [PubMed]


