
Fumarate hydratase deficiency induces chronic myeloid leukemia progression
Introduction
Chronic myeloid leukemia (CML) is a hematological malignancy characterized by the t(9;22) chromosomal translocation that results in the formation of the BCR-ABL1 fusion gene (1). The oncogene BCR-ABL1 produces the p210 fusion protein, which has tyrosine kinase activity (2). CML has two clinical phases: chronic phase (CP) is characterized by the expansion of myeloid progenitor cells with apparently normal differentiation and eventually followed by progression to acute leukemia called the blast phase (BP) (3). Although tyrosine kinase inhibitors (TKIs) have profoundly improved patient prognosis, the treatment of the BP is still difficult. The mechanism of progression to the BP is unclear.
The Warburg effect indicates that most tumor cells utilize glycolysis to proliferate (4,5). A previous study showed that in CML patients, TKI resistance was related to glycolysis. Zhu et al. found that PFKFB3, which controls the limiting step of glycolysis, is strongly associated with TKI resistance (6). In imatinib-resistant CML cells, the induction of HIF-1α expression at both the mRNA and protein levels is observed under normoxic conditions. The expression of target genes such as VEGFA, PGK1, and PKM2 is also elevated (7). Our previous study indicated that ATP generation by glycolysis is the main energy source in leukemia cells. BP cells had a high glycolytic status. We also found that fumarate expression is increased in CML-BP (8). There are two reasons for fumarate accumulation in cancer cells: biochemical inhibition and FH expression knockdown mediated by siRNA (9,10). Therefore, the fumarate accumulation in CML-BP may be because of the low activity of FH. We hypothesized that FH inhibition promotes CML progression by increasing glycolysis.
CML blast crisis is considered to be the consequence of alterations in DNA (11). However, the mechanism by which these alterations occur is less well understood. The high levels of ROS in CML cells increase DNA damage and the frequency of genetic mutations, and the DNA damage repair process is unfaithful. This repair is characterized by high frequency with large knockdowns and eventually results in disease progression (12-14). FH plays a key role in DNA damage repair. DNA damage induces FH to translocate into the nucleus and produce fumarate. The phosphorylation of the histone H2AX is one of the earliest responses to DNA double-strand breaks (DSBs), and the phosphorylation of CHK2 should be activated upon DSB induction. In the absence of FH, the phosphorylation of histone H2AX is impaired. The phosphorylation of CHK2 in FH-deficient cells is also much lower than that in control cells. Thus, FH deficiency may influence the recognition of and recovery from DNA damage (15).
In this study, we showed that FH expression was low in CML blast crisis samples. Knocking down FH expression induced glycolysis and enhanced the invasiveness of K562 cells. In addition, FH also influenced the sensitivity to DNA damage in K562 cells. Our findings revealed a new pathway of CML progression and may provide a novel approach for the treatment of CML-BP.
Methods
Patients and specimens
Bone marrow (BM) specimens were collected from 32 patients who had been diagnosed with CML at the Hematology Department of the First Affiliated Hospital of Nanjing Medical University (Nanjing, China) according to the 2008 criteria of the World Health Organization (WHO). Leukocytes were isolated from these specimens with Ficoll gradients and stored as frozen aliquots. All specimens were collected before chemotherapy. This study was approved by the institutional review board of the First Affiliated Hospital of Nanjing Medical University and conducted in accordance with the Declaration of Helsinki. Informed consent was obtained from all participants included in the study.
Cell culture
HL-60, K562, Ku812, and Jurkat cells were obtained from ATCC. Cell lines were grown in RPMI 1640 medium (Gibco BRL, USA) containing 10% FBS (HyClone). Cells were treated with 400 µm cobalt chloride (CoCl2) (Amresco, USA) or with 1 mg/mL 2-deoxy-D-glucose (2-DG) (Sigma-Aldrich, USA) for 48 h to induce or inhibit glycolysis, respectively. To determine the expression and localization of FH after DNA damage, K562 cells were inoculated in a 6-well plate (5×105 cells/well) and placed in a 37 °C cell incubator containing 5% CO2 for 18–24 h. Then, the cells were divided into five groups including K562 cells without hydroxyurea (Hu, Aladdin) and K562 cells treated with 1 mmol/L Hu for 1 h, 3 h, 7 h or 24 h. To verify the effect of FH on the relevant pathways after DNA damage, we created the following groups: K562 pcDNA3.1/FH and K562 pSilencer2.1/shR-Fh treated with 0, 0.25, 0.35, or 0.5 mmol/L Hu.
RNA extraction and quantitative reverse transcription-PCR
Total RNA was isolated using TRIzol (Invitrogen). The RNA isolation was performed according to the manufacturer’s protocols. Reverse transcription of the RNA was performed using an M-MLV Reverse transcription kit (Takara). Real-time qPCR was performed using SYBR Premix Ex Taq (Takara) with a Step One Plus real-time PCR system (Life Technology). The results were analyzed using Expression Suite software (Life Technology) and StepOne 2.3 software. All data were normalized using endogenous β-actin as the control, and the 2−ΔΔCt method was used to calculate the relative gene expression. Primers were purchased from Invitrogen and are listed in Table 1.
Table 1
| Gene | Primer sequence (5'→3') |
|---|---|
| FH | GAATCCAGGCCAATACAG |
| GTAAATCACTTTGGACCCAG | |
| HIF-1α | GCACAGGCCACATTCACG |
| TGAAGATTCAACCGGTTTAAGGA | |
| β-actin | CGTGACATTAAGGAGAAGCTG |
| CTAGAAGCATTTGCGGTGGAC | |
| HK2 | CAAAGTGACAGTGGGTGTGG |
| GCCAGGTCCTTCACTGTCTC | |
| LDHA | CTGCACCCAGATTTAGGGAC |
| CAACCTCCACCTAGAATCCTG | |
| PKM2 | CACCTGTACCGTGGCATCTTC |
| GTCAGCACAATGACCACATCTC | |
| PFK2 | ACCTAACCCGCTCATGAG |
| AATGGAATGGAACCGACAC | |
| FH-BamH I | CGCGGATCCGCCACCATGTACCGAGCACTTCGGC |
| FH-EcoR I | CCGGAATTCTCACTTTGGACCCAGCATG |
| HIF-1α-BamH I | CGCGGATCCGCCACCATGGAGGGCGCCGGCGGCG |
| HIF-1α-Xba I | GCTCTAGATCAGTTAACTTGATCCAAAGCTCTG |
| FH-shRNA-Top | GATCCGCTGCAATAGAAGTTCATGAACTCGAGTTCATGAACTTCTATTGCAGCTTTTTGA |
| FH-shRNA-Bot | AGCTTCAAAAAGCTGCAATAGAAGTTCATGAACTCGAGTTCATGAACTTCTATTGCAGCG |
| HIF-1α-shRNA-Top | GATCCGCTGAGGAAGAACTAAATCCAAACTCGAGTTTGGATTTAGTTCTTCCTCAGCTTTTTGA |
| HIF-1α-shRNA-Bot | AGCTTCAAAAAGCTGAGGAAGAACTAAATCCAAACTCGAGTTTGGATTTAGTTCTTCCTCAGCG |
Western blotting
The cellular protein content was extracted by RIPA buffer with a protease inhibitor cocktail. Lysates were spun at 16,000 g and 4 °C for 30 min and normalized by protein concentration. The lysates were separated by SDS-PAGE and electrotransferred to nitrocellulose membranes (Invitrogen). The membranes were blocked in PBS containing 0.1% (vol/vol) Tween-20 (PBS-T) and 4% (wt/vol) nonfat dry milk (Bio-Rad) for 1 h on a shaker at room temperature. Primary antibodies were added (anti-Fumarase/FH mouse monoclonal antibody, anti-HIF-1α mouse monoclonal antibody, anti-HK2, anti-PKM2, anti-PFK2, anti-LDHA, anti-β-actin, anti-p-H2AX and anti-p-Chk2 antibodies) (EnoGene, USA) and incubated overnight on a rocker at 4 °C. Secondary antibodies were diluted 1:2,000 in blocking buffer and incubated for one hour at room temperature. Images were acquired using LabWorksTM (UVP, USA).
Enzyme activity detection
After the cells grew to a density of 5×106/mL, the cell culture medium was absorbed and discarded, and the cultured cells were collected and washed with PBS. When the activities of HK2 and PFK2 were measured, 400 µL of extracting solution was added per every 2×106 cells, while when LDHA and PKM2 activities were measured, 500 µL of PBS was added. After the cells were lysed with ultrasonic sonication and centrifuged at 8,000 ×g for 10 min at 4 °C, the supernatant was collected. The protein content was measured using a BCA kit (Pierce) following the manufacturer’s instructions. Enzyme activity was measured by colorimetry following the manufacturer’s instructions.
ROS detection
Hu was added to K562 cells in 6-well plates. Then, the cells were incubated at 37 °C for 30 min with 500 µL of 10 µmol/L DCFH-DA probe (Beyotime Institute of Biotechnology, Shanghai, China), with shaking every 5 min. The cells were then washed with PBS (three times, 5 min each) to remove any remaining extracellular DCFH-DA probe. The fluorescence intensity, which represents the cellular ROS levels, was detected by flow cytometry (BD Biosciences).
Immunofluorescence experiments
Ionizing radiation-treated K562 cells were washed three times in PBS, fixed with 4% formaldehyde solution at 4 °C for 20 min and then washed with PBS three times. Then, the cells were incubated with 0.5% Triton X-100 at 4 °C for 5 min and incubated with a blocking solution (10% donkey serum in PBS) for 2 h at room temperature after three PBS washes. The cells were then incubated with primary antibodies against p-H2AX (EnoGene, USA) diluted in the blocking solution and incubated in a wet box overnight at 4 °C. After three washes with PBS, the cells were incubated with a secondary antibody for 2 h at room temperature. Then, the cells were incubated with DAPI (1:1,000, diluted with PBS) for 5 min at 4 °C. Images were acquired with an Olympus fluorescence microscope after three washes with PBS.
shRNA knockdown and overexpression vector construction
We designed and synthesized shRNAs specific for FH and HIF-1α based on Ambion’s software (Table 1). An FH-shRNA cell line (pSilencer2.1-U6/shR-FH) and HIF-1α-shRNA cell line (pSilencer2.1-U6/shR-HIF-1α) were constructed by lipofectin-mediated gene transfer. PCR was used to amplify fragments containing FH and HIF-1α. The primers were FH-BamHI, FH-EcoRI, HIF-1α-BamHI and HIF-1α-XbaI (Table 1). The BamHI, EcoRI and XbaI sites were cut off, and the fragments were connected to the pcDNA 3.1 plasmid (EnoGene, USA) by a ligase (T4 DNA ligase, Promega, the United States). The overexpression model was obtained by Lipofectamine 2000 Reagent (Invitrogen) transfection, limiting dilution cloning and cell proliferation.
Flow cytometry
For apoptosis staining, 10 µL of Annexin-V-R-PE (ApoScreen Annexin V Apoptosis Kit, Southern Biotech, USA) was added to 5×105 cells after the last wash and incubated for 20 min in the dark in an ice bath. The samples were washed once, and 380 µL of 1× binding buffer and 2 µL of 7-aminoactinomycin D (7AAD) (eBioscience) were added within 4 h before FACS analysis. For proliferation detection, 0.5–1 mL of PI (100 mg/L) (Propidium iodide, Roche, Switzerland) was added to each sample, fully mixed and incubated at room temperature for 30 min in the dark. FACS analysis was performed immediately using a flow cytometer (BD Biosciences).
Invasiveness detection
Cell invasion was assayed by a Transwell (Corning Incorporated) with Matrigel (BD Biosciences, USA). Each Transwell chamber was coated with 100 µL of Matrigel and placed into a 24-well plate for 30 min at 37 °C. A total of 300 µL of cell suspension (105/100 µL) in serum-free RPMI 1640 medium was added to the upper chamber. In the experimental group, 400 µmol/L CoCl2 or 1 mg/mL 2-DG was added to the upper chamber, while in the control group, PBS was added. After 48 h of coculture, the cells attached to the upper and lower surfaces of the filter membrane were fixed with 4% paraformaldehyde for 20 min. The cells on the upper surface of the filter membrane were removed with a swab, and the membrane was washed with PBS. Then, the filter membrane was stained with 0.1% crystal violet and washed in PBS three times. Cell images were obtained after the filter membrane was dry.
Statistics
Statistical analyses were performed using Student’s t-test with SPSS version 22.0 (IBM, USA) and GraphPad Prism version 5.01 for Windows (GraphPad Software). A P value less than 0.05 was considered statistically significant.
Results
FH, HIF-1α, HK2, PKM2, LDHA and PFK2 expression in CML-CP and CML-BP patients
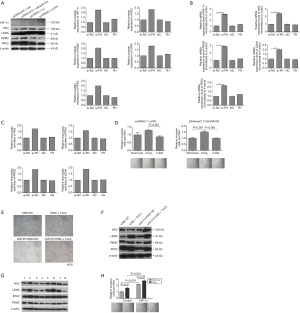
We detected the protein levels of FH, HIF-1α, HK2, PKM2, LDHA and PFK2 in bone marrow mononuclear cells of 11 blast crisis patients and 21 CP patients. FH expression was significantly lower in the CML-BP patients than in the CML-CP patients (Figure 1A, P=0.025). The expression of HIF-1α and the glycolytic enzymes HK2, LDHA and PFK2 was increased in CML-BP (Figure 1A). Therefore, the CML-BP patients had low FH expression and high glycolysis levels.
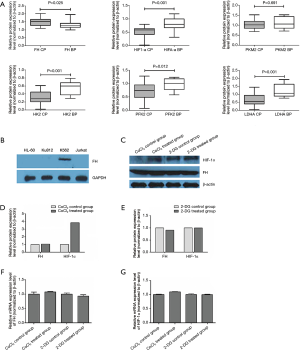
FH expressed in K562 cell line and glycolysis induction accompanied the expression of HIF-1α
Western blot was used to test the expression of FH in four human leukemia cell lines: HL-60, K562, KU812, and Jurkat. Among these cell lines, only the K562 cell line, which was derived from a CML patient in blast crisis, exhibited FH expression (Figure 1B). CoCl2 was used to induce glycolysis, and 2-DG was used to block glycolysis in K562 cells. We detected the mRNA and protein expression of FH and HIF-1α after treatment with CoCl2 or 2-DG. There was no significant difference in FH expression before and after induction (Figure 1C,D,E,F). CoCl2 stimulated the development of a hypoxic environment and increased the protein expression of HIF-1α (Figure 1C,D). The HIF-1α mRNA transcript was expressed constitutively in K562 cells under both normal and hypoxic conditions, and this expression was not induced by CoCl2 (Figure 1G).
Knocking down FH expression induced HIF-1α expression and elevated the glycolysis state, increasing the invasiveness of K562 cells
To test whether FH could regulate glycolysis in K562 cells, we detected the expression of glycolytic enzymes, including HK2, PKM2, LDHA and PFK2, and enzyme activity was also tested by colorimetry. After knocking down FH expression, glycolytic enzyme expression was increased (Figure S1A,B). HIF-1α is an important regulator of glycolysis. To test whether FH regulates glycolysis through HIF-1α, we also detected the expression of HIF-1α. Knocking down FH expression increased HIF-1α expression at both the mRNA and protein levels (Figure S1A,B). The activities of glycolytic enzymes were increased after FH expression knockdown (Figure S1C). However, the overexpression of FH had no significant effect on HIF-1α or glycolytic enzyme expression. In addition, we verified the change in invasiveness in K562 cells after FH expression knockdown or overexpression at different levels of glycolysis (Figure S1D). Among the FH-overexpressing cells, the invasiveness of the CoCl2 group was higher than that of the 2-DG group. For the cells with knocked down FH expression, the invasiveness of the CoCl2 group was higher than that of both the normoxia and 2-DG groups (P<0.001 for both).
FH expression knockdown increased the levels of glycolysis and enhanced invasiveness of K562 cells through the HIF-1α pathway
We used CoCl2 to induce HIF-1α expression in the cells with knocked down FH expression. After induction, the cell volume decreased significantly, cytoplasm increased, and nucleoplasm ratio decreased (Figure S1E). As HIF-1α expression increased, the protein expression of the glycolytic enzymes increased in the FH expression knockdown and control groups (Figure S1F). The synergistic effect of HIF-1α expression and FH expression knockdown on glycolysis induction was significant.
Next, we tested the effect of knocking down FH expression combined HIF-1α overexpression or knockdown on glycolysis. We found that glycolytic enzyme expression was increased in the HIF-1α overexpression group, while it was decreased in the HIF-1α expression knockdown group (Figure S1G). FH expression knockdown increased glycolytic enzyme expression. Furthermore, knocking down FH expression combined with overexpressing HIF-1α significantly increased glycolytic enzyme expression (Figure S1G). However, knocking down FH and HIF-1α expression slightly decreased the level of glycolysis, which indicated that FH may regulate glycolysis through the HIF-1α pathway. At the same time, we detected the invasiveness of each group. HIF-1α overexpression enhanced the invasiveness of FH expression-knockdown K562 cells. In the hypoxic state induced by CoCl2, invasiveness was increased significantly (Figure S1H).
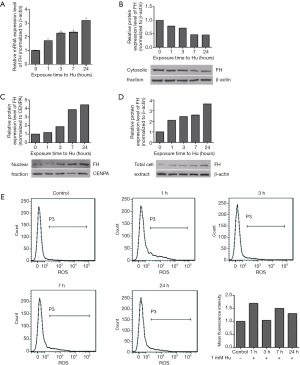
DNA damage promoted FH translocation to the nucleus in K562 cells
Hu (1 mmol/L) was used to induce DNA damage. Over time, the mRNA and protein expression of FH increased (Figure 2). At the same time, FH translocated to the nucleus, and this movement was reflected in the decreased cytoplasmic FH protein level and increased nuclear FH protein level (Figure 2B,C). Intracellular ROS levels were also measured by flow cytometry. The ROS levels first increased and then decreased, increasing to a maximum at 1 h (Figure 2E).
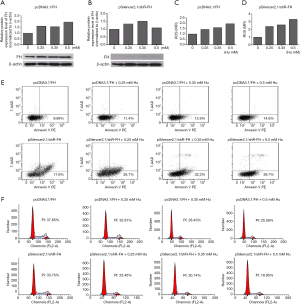
FH was required in DNA damage repair, and FH expression knockdown decreased the DNA repair capacity of K562 cells
After DNA damage was induced by Hu at different concentrations ranging from 0 to 0.5 mmol/L, FH protein expression was increased in the FH overexpression group (Figure 3A). However, in the FH expression knockdown group, FH expression first increased and then decreased (Figure 3B). The ROS level in the FH overexpression group showed an increasing trend (Figure 3C), while that in the FH expression knockdown group was significantly increased (Figure 3D). The FH overexpression group had a slightly increased apoptosis rate (from 9.88% to 14.6%) and a decreased proliferation rate (from 37.65% to 25.58%). The FH expression knockdown group showed a significantly increased apoptosis rate (from 17.6% to 39.7%) and a decreased proliferation rate (from 33.75% to 18.95%) (Figure 3E,F).
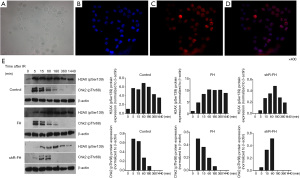
Peaks in DNA damage biomarker levels were later in K562 cells with knocked down FH expression
To detect the effect of FH on DNA damage repair, we used 5 Gy γ-ionizing radiation (IR) to cause DNA damage in K562 cells. We detected the expression of phosphorylated H2AX (γH2AX) and Chk2 activation (p-Chk2), which are two biomarkers of DNA damage and repair. Immunofluorescence staining showed that the expression of phosphorylated H2AX was mainly located in the nucleus (Figure 4A,B,C,D). The protein expression results showed that the γH2AX level in the control group increased rapidly from the beginning of detection, reached a peak at 1 h, and then decreased rapidly. In the FH overexpression group, γH2AX expression began to increase after irradiation, reached a peak at 1 h, and then gradually decreased. The γH2AX expression in the FH expression knockdown group increased at 5 min after irradiation, peaked at 3 h, and then decreased (Figure 4E). The p-Chk2 expression of the K562 cells in the control group increased and peaked at 5 min after irradiation. The p-Chk2 expression in the overexpression group began to increase at 5 min after irradiation and could not be detected after 3 h. In the FH knockdown group, p-Chk2 expression increased 5 min after irradiation, reached a peak at 1 h, and could not be detected later (Figure 4E).
Discussion
Fumarate hydratase (FH) is a TCA enzyme that catalyzes the reversible hydration of fumarate to malate. It has been reported that FH loss and fumarate accumulation are related to the aggressive features of hereditary leiomyomatosis and renal cell carcinoma (HLRCC) (16). Fumarate is one of the analogues of 2-oxoglutarate, and it can competitively inhibit several 2-oxoglutarate-dependent dioxygenases (2-OGDDs), including HIF prolyl hydroxylases (PHDs) and ten-eleven-translocation 5-methylcytosine dioxygenases (TETs) (17). It has been identified that the stabilization of the hypoxia-inducible factor HIF-1α in HLRCC is because of the inhibition of PHDs (18). HIF-1α stabilization increases the transcription of its target genes GLUT1 and VEGF, which can promote the vascularization and glucose transport needed for tumor growth (19).
Our previous study indicated that CML-BP cells proliferate with glycolysis as their energy source and that the level of fumarate is higher in CML-BP than in CML-CP (8). Therefore, we wanted to determine whether the increased level of glycolysis is the reason for disease progression. Our findings showed that knocking down FH expression increased the expression and activity of glycolytic enzymes. Importantly, FH expression knockdown also enhanced the invasiveness of K562 cells. Furthermore, we wanted to determine whether FH regulates glycolysis through the HIF-1α pathway. We found that FH expression knockdown increased the expression of HIF-1α. When HIF-1α expression was knocked down, glycolysis was significantly inhibited, and even FH expression knockdown could not reverse this inhibition. In addition, FH expression knockdown cooperated with HIF-1α in glycolysis regulation.
Genomic instability is believed to be responsible for CML blast crisis. The high levels of ROS created in BCR-ABL cells cause chronic oxidative DNA damage and result in DNA DSBs. DSBs are the main DNA damage in CML cells, and the main mechanism to repair the damage is the alternative nonhomologous end-joining pathway (ALT NHEJ). Although BCR-ABL enhances repair activity, the repair of DSBs in Ph+ CML cells is unfaithful (12). The NHEJ repair pathway is DNA-dependent protein kinase (DNA-PK) dependent in mammalian cells. However, in BCR-ABL cells, the activity of the DNA PK-dependent NHEJ pathway is decreased, and the activity of the ALT NHEJ pathway, which is an error-prone repair process characterized by a high frequency of large deletions, is increased. The decreased activity of the DNA PK-dependent NHEJ pathway may be due to the elevated ROS levels. High levels of the ALT NHEJ repair-related enzymes DNA ligase IIIα and poly-(ADP-ribose) polymerase 1 (PARP1) are the main evidence (13,14,20).
We used 1 mmol/L Hu to induce DNA damage in K562 cells and found that FH expression was increased and cytoplasmic FH moved into the nucleus. The ROS level first increased and then decreased, which may indicate that following the increase in FH expression, DNA damage repair was underway. However, with FH expression knockdown, the ROS level was increased obviously and did not exhibit a downward trend. The phosphorylation of H2AX is the earliest response to DSBs. In FH expression-knockdown K562 cells, the peak levels of phosphorylated H2AX and the cell cycle checkpoint Chk2 occurred later than in the control cells. These results indicated that FH expression knockdown may influence the recognition and recovery from DNA damage.
Conclusions
In conclusion, our results showed that a functional deficiency in FH may induce the Warburg effect through the HIF-1α pathway and lead to CML blast crisis. FH deficiency also influenced DNA damage repair in K562 cells. Therefore, our findings revealed a novel pathway of CML progression, and HIF-1α may be a powerful target for future clinical use in CML-BP.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.03.23). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the institutional review board of the First Affiliated Hospital of Nanjing Medical University (IRB number: 2011-SRFA-033). Informed consent was obtained from all individual participants included in the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Spiers AS. Clinical manifestations of chronic granulocytic leukemia. Semin Oncol 1995;22:380-95. [PubMed]
- Gorska-Flipot I, Norman C, Addy L, et al. Molecular pathology of chronic myelogenous leukemia. Tumour Biol 1990;11:25-43. [Crossref] [PubMed]
- Maru Y. Molecular biology of chronic myeloid leukemia. Cancer Sci 2012;103:1601-10. [Crossref] [PubMed]
- Warburg O. On the origin of cancer cells. Science 1956;123:309-14. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Zhu Y, Lu L, Qiao C, et al. Targeting PFKFB3 sensitizes chronic myelogenous leukemia cells to tyrosine kinase inhibitor. Oncogene 2018;37:2837-49. [Crossref] [PubMed]
- Zhao F, Mancuso A, Bui TV, et al. Imatinib resistance associated with BCR-ABL upregulation is dependent on HIF-1alpha-induced metabolic reprograming. Oncogene 2010;29:2962-72. [Crossref] [PubMed]
- A J, Qian S, Wang G, et al. Chronic myeloid leukemia patients sensitive and resistant to imatinib treatment show different metabolic responses. PLoS One 2010;5:e13186.
- Maxwell PH. Seeing the smoking gun: a sensitive and specific method to visualize loss of the tumour suppressor, fumarate hydratase, in human tissues. J Pathol 2011;225:1-3. [Crossref] [PubMed]
- Ashrafian H, O'Flaherty L, Adam J, et al. Expression profiling in progressive stages of fumarate-hydratase deficiency: the contribution of metabolic changes to tumorigenesis. Cancer Res 2010;70:9153-65. [Crossref] [PubMed]
- Koschmieder S, Vetrie D. Epigenetic dysregulation in chronic myeloid leukaemia: A myriad of mechanisms and therapeutic options. Semin Cancer Biol 2018;51:180-97. [Crossref] [PubMed]
- Nowicki MO, Falinski R, Koptyra M, et al. BCR/ABL oncogenic kinase promotes unfaithful repair of the reactive oxygen species-dependent DNA double-strand breaks. Blood 2004;104:3746-53. [Crossref] [PubMed]
- Sallmyr A, Fan J, Rassool FV. Genomic instability in myeloid malignancies: increased reactive oxygen species (ROS), DNA double strand breaks (DSBs) and error-prone repair. Cancer Lett 2008;270:1-9. [Crossref] [PubMed]
- Tobin LA, Robert C, Rapoport AP, et al. Targeting abnormal DNA double-strand break repair in tyrosine kinase inhibitor-resistant chronic myeloid leukemias. Oncogene 2013;32:1784-93. [Crossref] [PubMed]
- Yogev O, Yogev O, Singer E, et al. Fumarase: a mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol 2010;8:e1000328. [Crossref] [PubMed]
- Bhola PT, Gilpin C, Smith A, et al. A retrospective review of 48 individuals, including 12 families, molecularly diagnosed with hereditary leiomyomatosis and renal cell cancer (HLRCC). Fam Cancer 2018;17:615-20. [Crossref] [PubMed]
- Yum S, Choi J, Hong S, et al. Hyperoxia attenuates the inhibitory effect of nitric oxide donors on HIF prolyl-4-hydroxylase-2: Implication on discriminative effect of nitric oxide on HIF prolyl-4-hydroxylase-2 and collagen prolyl-4-hydroxylase. Biochem Pharmacol 2011;82:485-90. [Crossref] [PubMed]
- Selak MA, Armour SM, MacKenzie ED, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell 2005;7:77-85. [Crossref] [PubMed]
- Isaacs JS, Jung YJ, Mole DR, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell 2005;8:143-53. [Crossref] [PubMed]
- Sallmyr A, Tomkinson AE, Rassool FV. Up-regulation of WRN and DNA ligase IIIalpha in chronic myeloid leukemia: consequences for the repair of DNA double-strand breaks. Blood 2008;112:1413-23. [Crossref] [PubMed]


