Identification of RRAS gene related to nasopharyngeal carcinoma based on pathway and network-based analyses
Introduction
Nasopharyngeal carcinoma (NPC), which occurs primarily in the nasopharynx epithelium, is mainly distributed in the eastern and southeastern parts of Asia, south-central Asia, and northern and eastern Africa (1,2). As a highly aggressive neoplasm, the prognosis of NPC patients remains poor, and approximately 15% of patients develop distant metastasis (2).
Genetic predisposition, salted fish consumption, and Epstein-Barr virus (EBV) infection have been proven to be important risk factors for NPC (1,3). Although some biomarkers and signaling pathways have been identified in NPC, it is necessary to conduct a comprehensive analysis of genes that play an important role in NPC carcinogenesis.
RRAS is a Ras-related GTPase (4,5). The human RRAS gene is located on chromosome 19q13.3 and was detected by low-stringency hybridization using a v-H-ras probe (6). RRAS was reported to be involved in several cell functions, including the enhancement of integrin function, regulation of cell adhesion, invasion and migration (7-9). The role of RRAS has been reported in breast cancer, colorectal cancer, and melanoma, but it has not been studied in nasopharyngeal cancer (6,7,10).
In this study, we constructed an NPC-related gene set (NPCgset). Kyoto Encyclopedia of Genes and Genomes (KEGG) and pathway crosstalk analyses were used to reveal the signaling pathway modules in NPC. Then, we constructed two protein-protein interaction (PPI) networks to find new genes associated with NPC. Combined with the results of experiments using NPC cell lines and clinical samples, we found that RRAS was down-regulated in NPC and might be a new potential biomarker.
Methods
Identification of NPC-related genes
The online database PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) was retrieved by two independent researchers with the following terms: ("Nasopharyngeal carcinoma") and ("Genotype"[Mesh] OR "Genetic Association Studies"[Mesh]). Next, we reviewed these publications and narrowed the scope of the study by the following criteria: (I) biological experiments must validate genes associated with NPC or have clinical significance; (II) non-coding genes were excluded; (III) genes predicted by bioinformatics methods alone were excluded; (IV) the studies reporting insignificant associations were excluded. The selected genes were used for bioinformatics analysis.
Enrichment analysis for NPC-related genes
GO, and KEGG pathway enrichment analysis was completed by WebGestalt (http://www.webgestalt.org/option.php), which is an online tool that incorporates information from different resources to perform multiple analysis such as GO and KEGG on a given gene list. For both analysis, FDR <0.05 was considered to be significant.
Pathway crosstalk analysis
The candidate pathways obtained from KEGG analysis were used for pathway crosstalk analysis. We introduced two parameters to explore the interaction between each pair of pathways, i.e., the Jaccard Coefficient (JC) = |(A∩B)/(A∪B)| and the Overlap Coefficient (OC) = |A∩B|/min(|A|,|B|). The average scores of the JC and OC were calculated as grades to rank the pathway pairs. Pathways containing less than ten genes were not included. Pathway pairs with mean values of JC and OC less than 0.5 were removed. The results of pathway crosstalk analysis were visualized using the software Cytoscape.
PPI network construction
The PPI network of candidate genes was constructed using the STRING database (http://string-db.org), and the combined score >0.4 was used as the cut-off criterion. Cytoscape was used to visualize the PPI network, and the hub genes of the network were selected according to the degree that calculated by plugin CytoHubba.
Patients and samples
Thirty-one cases of fresh tissues, including 16 NPC and 15 nasopharyngitis tissues were collected from Sun Yat-sen University Cancer Centre (SYSUCC), and total RNA was extracted for the qRT-PCR assay. Paraffin-embedded specimens from 65 primary NPC patients, including 50 males (77%) and 15 females (23%), were collected for the immunohistochemistry (IHC) assay. The median age of the patients was 45 years with a range from 10 to 66. None of the patients had received radiotherapy or chemotherapy before sampling. The Research Ethics Committee of SYSUCC approved our research, and all patients signed informed consent forms.
Cell lines and transfections
The human NPC cell lines 5-8F, 6-10B and HK1 were maintained in RPMI-1640 medium (Gibco, Thermo Fisher Scientific, China), C666-1 cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, China), and they were all supplemented with 10% fetal bovine serum (Gibco, Thermo Fisher Scientific, South America). NP69, an immortalized nasopharyngeal epithelium cell line, was grown in defined K-SFM medium supplemented with EGF and bovine pituitary extract (Gibco, Thermo Fisher Scientific, USA). These cell lines were cultured in an atmosphere of 5% CO2 at 37 °C. siRNA duplexes against RRAS and the negative control (NC) were purchased from GenePharma (Shanghai, China). Cell transfection was performed using Lipofectamine 3000 (Invitrogen, USA) as described previously (11).
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted with TRIzol (Invitrogen, USA) following the manufacturer's protocol as described previously (11). The primer sequences for the qRT-PCR assays were as follows: RRAS sense: 5'-TGTCTGACTACGACCCCACT-3'; RRAS anti-sense: 5'-GCCCACCTCGTTGAAACTCT-3'; IRS2 sense: 5'-GCCACCATCGTGAAAGAGTGA-3'; IRS2 anti-sense: 5'-CCATCCGGGAACAAGGGAAA-3'; GAPDH sense: 5'-CTCCTCCTGTTCGACAGTCAGC-3'; GAPDH anti-sense: 5'-CCCAATACGACCAAATCCGTT-3'.
IHC staining
The IHC assay was performed using GTVision III Detection System/Mo &Rb [Gene Tech (Shanghai) Co., Ltd., China], following the manufacturer's instruction, and the results were scored by two evaluators independently. The percentage of positive cells was graded as 0 (≤10%), 1 (11–25%), 2 (26–50%), 3 (51–75%), and 4 (>75%), and the intensities were graded as negative [0], weakly positive [1], moderately positive [2] and strongly positive [3]. The multiplied intensity score and percentage score was used as the total IHC score.
Western blotting
Proteins were harvested from cell lines by the Whole Cell Lysis Assay Kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) according to the manufacturer’s instructions. The concentration of total protein was quantified by the BCA assay kit (Beyotime Institute of Biotechnology) and Gen5 Software (CHS 2.06; BioTek Instruments, Winooski, VT, USA). 10% SDS-PAGE and PVDF membranes (GE Healthcare Life Sciences, UK) were used to separate and transfer the protein. Next, the membranes were incubated with primary antibodies, including E-cadherin (1:1,000; CST; cat. no. 3195), N-cadherin (1:1,000; CST; cat. no. 13116), Vimentin (1:1,000; CST; cat. no. 5741), RRAS (Proteintech Group Inc.; cat. no. 27457-1-AP) and GAPDH (1:1,000; CST; cat. no. 5174) at 4 °C overnight. Secondary antibodies (HRP-linked anti-rabbit IgG; 1:5,000; CST, cat. no. 7074) were subsequently incubated with the membranes for 1 hour at room temperature. The protein signals were detected using an electrochemiluminescence (ECL) system (Tanon, China).
CCK8 proliferation assay
The CCK8 Cell Counting Kit (JingXin Biological Technology, Guangzhou, China) was used to detect cell proliferation according to the manufacturer’s instructions. Absorbance at 450 nm was measured using a microplate reader (SpectraMax® M5 Multi-Mode Microplate Reader; Molecular Devices LLC, Sunnyvale, CA, USA).
Colony formation assays
HK1 cells with different treatments were seeded in six-well plates (1,000 cells per well) and were cultured for two weeks. Colonies were fixed with methanol for 10 min and then were stained with 0.5% crystal violet (Weijia Biology Science and Technology Co., Ltd) for 10 min. The colonies were then counted under a dissection microscope.
Invasion assays
Cells were counted and diluted with serum-free RPMI-1640 medium to 5×104 cells/200 µL. Next, the cell suspensions were added to Matrigel-coated chambers (BD Biosciences), which were inserted into a 24-well plate with 10% FBS in RPMI-1640 medium (700 µL/well). The chambers were removed 36 hours later, and the cells left in the upper chambers were swabbed. The invasive cells in five random fields per well were counted under the microscope.
Statistical analysis
Statistical analysis was conducted using SPSS software version 19.0 (IBM Corp., Armonk, NY, USA). Repeated measures ANOVA were used to analyze the results of CCK8. Student’s t-test tested the differences between the two groups in other cell function experiments. Kaplan-Meier analysis and the log-rank test were used to explore the relationship between RRAS expression and overall survival (OS) or progression-free survival (PFS). χ2-test analyzed the correlation between RRAS expression and clinicopathological features of NPC patients. P values less than 0.05 were considered to indicate statistical significance. The Cox regression analysis was used to analyze the association between the expression of RRAS and OS after adjusting for known important clinicopathologic features, and P values less than 0.1 were considered to indicate statistical significance.
Results
Identification of genes associated with NPC
By October 10, 2018, 2,799 publications were retrieved in the PubMed database, and 531 of them were selected after a review of the abstracts. We reviewed these publications for details and an NPC-related gene set (NPCgset) with 552 members from 514 reports was constructed. The details of all genes in the gene set are provided in http://fp.amegroups.cn/cms/tcr.2019.04.04-1.pdf.
Gene Ontology (GO) and pathway enrichment analysis in the NPCgset
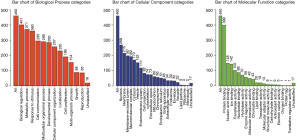
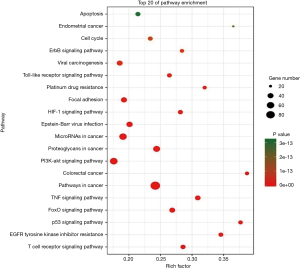
Our NPCgset contained 552 user IDs, in which 431 were annotated to the selected GO functional categories and in the reference gene list, and these genes were used for GO analysis (Figure 1). The details of the top 39 GO terms are shown in Table S1. GO terms related to cell proliferation (e.g., epithelial cell proliferation and positive regulation of cell cycle), and migration (e.g., cell-substrate adhesion and regulation of cell-cell adhesion) have been shown to play an important role in the progression of NPC. In the KEGG pathway enrichment analysis, 127 significantly enrichment pathways were found. The top 20 pathways are shown in Figure 2. Among them, EGFR tyrosine kinase inhibitor resistance and EBV infection deserve particular attention, because EGFR-targeted therapy has been introduced in NPC treatment (12) and EBV infection is a special risk factor for NPC (13). Other pathways, including tumor-associated biological processes (e.g., p53 signaling pathway, microRNAs in cancer, and pathways in cancer) and pathways related to the immune response (e.g., T-cell receptor signaling pathway and Toll-like receptor signaling pathway) were also significantly enriched in the NPCgset. These results suggested that our NPCgset is relatively reliable and is competent for the following studies.
Pathway crosstalk analysis
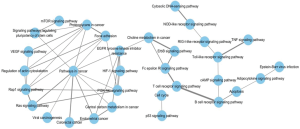
To remove the interference of redundant information and make a more concise understanding of the interactions between the pathways extracted from enrichment analysis, we introduced pathway crosstalk analysis, which has been reported previously (14). The theoretical basis of the method was that if two pathways shared a proportion of a gene set, they were considered to have crosstalk (15). Among the pathways enriched in KEGG analysis, 51 pathways were closely related to NPC according to previous studies (http://fp.amegroups.cn/cms/tcr.2019.04.04-2.pdf), of which 32 met our criteria and were used to build the network. According to our crosstalk, two representative models were identified, and pathways involved in each model shared more common features (Figure 3). One module was mainly related to the carcinogenesis process, including pathways in cancer, proteoglycans in cancer and VEGF signaling pathway. The other module contained the Toll-like receptor signaling pathway, T-cell receptor signaling pathway and so on, which presented an obvious correlation with the immune system. The interaction of two pathways, EGFR tyrosine kinase inhibitor resistance and choline metabolism in cancer, acted as the main connection between the two models.
PPI analysis
Since EBV infection and EGFR tyrosine kinase inhibitor resistance has been proven to be important in the treatment of NPC, our study first focused on these two pathways. Eight duplicate genes in the two pathways were extracted, i.e., AKT3, AKT1, GSK3B, PIK3CA, PIK3R1, PLCG1, BCL2, and STAT3. The PPI network of these eight genes was obtained using STRING (Figure 4A), and the connectivity degrees were calculated by CytoHubba (Table 1). Among them, PIK3CA and AKT1 with the highest degrees were chosen as hub genes. Proteins that interact with PIK3CA were predicted in STRING (Figure 4B), most of which were included in our NPCgset except RRAS and IRS2, which have not been reported in NPC. To confirm that the RRAS and IRS2 proteins interacted with PIK3CA play an important role in NPC carcinogenesis, we conducted the following experiment.
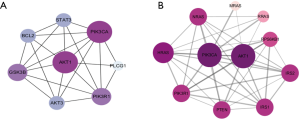
Table 1
| Gene symbol | Degree |
|---|---|
| PLCG1 | 3 |
| GSK3B | 6 |
| STAT3 | 5 |
| AKT3 | 5 |
| BCL2 | 5 |
| PIK3R1 | 6 |
| PIK3CA | 7 |
| AKT1 | 7 |
RRAS is down-regulated in NPC tissues and cells
The mRNA expression levels of RRAS and IRS2 were evaluated by qRT-PCR in NPC cell lines (HK1, C666, 5-8F, 6-10B), immortalized nasopharyngeal epithelial cell line NP69, fresh NPC tissues, and nasopharyngitis tissues. The results demonstrated that the expression of IRS2 mRNA showed no significant difference in NPC cell lines and tissues compared with the control (Figure S1). However, RRAS mRNA expression was lower in NPC cell lines (Figure 5A) and significantly decreased in NPC tissues compared with non-tumor tissues (Figure 5B). Moreover, the lower protein level of RRAS in NPC cell lines than in NP69 was also confirmed by western blotting (Figure 5C). Next, RRAS expression was evaluated in 65 paraffin-embedded NPC specimens by IHC assay. Positive expression of RRAS protein was detected on the cytoplasm and cell membrane. The IHC scores for RRAS in these NPC tissues ranged from 0 to 12, and the median score was 4 (Figure 5D). The median value of IHC score (≥4) was used as a cut-off value. Lower expression of RRAS was significantly related to advanced clinical stages (P=0.019) (Table 2). No significant differences were found between RRAS expression and other clinicopathological features. Higher expression of RRAS was associated with longer OS of patients (P=0.008) (Figure 5E). Multivariate analysis showed that the down-regulation of RRAS (RRAS expression: low vs. high; HR =2.244, 95% CI: 0.925–5.441, P=0.074) and distant metastasis (distant metastasis: no vs. yes; HR =0.146, 95% CI: 0.059–0.364, P<0.001) were significantly associated with a shorter OS (Table 3). Our results indicated that RRAS might be associated with the prognosis of NPC patients.
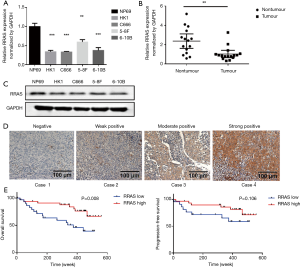
Table 2
| Variable | No. | RRAS expression | P value (χ2-test) | |
|---|---|---|---|---|
| Low (n=32) | High (n=33) | |||
| Gender | 0.341 | |||
| Female | 15 | 9 (60.0) | 6 (40.0) | |
| Male | 50 | 23 (46.0) | 27 (54.0) | |
| Age | 0.108 | |||
| <45 | 30 | 18 (60.0) | 12 (40.0) | |
| ≥45 | 35 | 14 (40.0) | 21 (60.0) | |
| EA/IgA | 0.388 | |||
| <1:20 | 31 | 17 (54.8) | 14 (45.2) | |
| ≥1:20 | 34 | 15 (44.1) | 19 (55.9) | |
| VCA/IgA | 0.953 | |||
| <1:160 | 12 | 6 (50.0) | 6 (50.0) | |
| ≥1:160 | 53 | 26 (49.1) | 27 (50.9) | |
| Clinical staging | 0.019* | |||
| I–III | 34 | 12 (35.3) | 22 (64.7) | |
| VI | 31 | 20 (64.5) | 11 (35.5) | |
| Distant metastasis | 0.082 | |||
| No | 47 | 20 (42.6) | 27 (57.4) | |
| Yes | 18 | 12 (66.7) | 6 (33.3) | |
*P<0.05. NPC, nasopharyngeal carcinoma.
Table 3
| Factors | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Age: <45 vs. ≥45 | 0.570 (0.240–1.353) | 0.203 | – | – | |
| Gender: female vs. male | 0.976 (0.361–2.641) | 0.962 | – | – | |
| EA/IgA: <1:20 vs. ≥1:20 | 0.928 (0.407–2.119) | 0.86 | – | – | |
| VCA/IgA: <1:160 vs. ≥1:160 | 0.899 (0.304–2.659) | 0.0847 | – | – | |
| Clinical staging: I–III vs. VI | 0.307 (0.129–0.727) | 0.007** | – | – | |
| Distant metastasis: no vs. yes | 0.126 (0.051–0.309) | <0.001*** | 0.146 (0.059–0.364) | <0.001*** | |
| RRAS expression: low vs. high | 3.045 (1.282–7.230) | 0.012* | 2.244 (0.925–5.441) | 0.074# | |
#, P<0.1; *, P<0.05; **, P<0.01; ***, P<0.001. NPC, nasopharyngeal carcinoma.
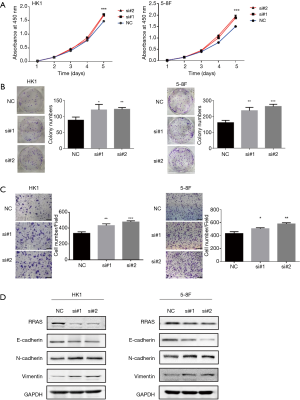
RRAS knockdown promotes cell proliferation and invasion
To explore the role of RRAS in NPC cells, RRAS mRNA knockdown was mediated by two specific siRNA oligos. RRAS knockdown significantly promoted the growth rate, colony formation and invasive ability of HK1 and 5-8F cells (Figure 6A,B,C). The depletion of RRAS was verified by western blotting (Figure 6D). Moreover, RRAS inhibition increased the expression of N-cadherin and Vimentin and decreased E-cadherin expression in NPC cells (Figure 6D). The results indicated that RRAS might suppress proliferation, invasion and EMT phenotype of NPC cells.
Discussion
In this study, we constructed a systematic framework of NPC-related biochemical processes by extracting the genes associated with NPC from publications and detecting the interaction of these genes using enrichment and network analyses. Then, we successfully predicted a new NPC related gene RRAS. The GO enrichment analysis of the NPCgset identified the biological processes related to NPC, and most of them were consistent with previous studies. For example, genes related to the cell-cell adhesion, epithelial cell proliferation, positive regulation of the cell cycle and kinase activity have been extensively studied in NPC (3,11,16). However, some new terms enriched in our analysis remains to be explored, such as the response to steroid hormone. The role of steroid hormones has been studied in several cancers, including breast cancer, prostate cancer and bladder cancer (17-19). Dai et al. (20) found that steroid hormone metabolomics can be a promising biomarker tool for early HCC. The NPC incidence rate in men are two to three times more than in women (1); in addition to the differences in living habits, the response to steroid hormone might be associated with this phenomenon.
To eliminate redundant information in pathway crosstalk analysis, pathways such as dilated cardiomyopathy and amoebiasis were excepted, and two main modules were constructed. One module was mainly related to carcinogenesis. The PI3K-Akt signaling pathway, MAPK signaling pathway, VEGF signaling pathway, and mTOR signaling pathway in this module has been reported to be important in NPC (21-24). The pathways in the other module were mainly associated with the immune response, suggesting that both innate immunity and specific immunity contribute to NPC.
We found that EGFR tyrosine kinase inhibitor resistance and choline metabolism in cancer act as a bridge between the two modules. EGFR was reported to be expressed in more than 85% of NPC patients (25) and predicts a worse prognosis (26). However, Hsu et al. (27) reported that the effect of inhibiting EGFR activation in NPC alone was not satisfactory. Therefore, it is of great value to explore the mechanism of EGFR inhibitor resistance. Next, we focused on EBV infection because it is perhaps one of the most extensively studied risk factors of NPC (24).
Two hub genes PIK3CA and AKT1 in PPI network were all proven to be associated with NPC (26). Because more studies on AKT1 have been reported in NPC, we focused on PIK3CA. Among the proteins that interact with PIK3CA, two of them (RRAS and IRS2) has not been reported in NPC until now. Others such as HRAS, NRAS, IRS1, PI3KR1 (P85), and S6K have been included in our NPCgset. MRAS was shown to bind to RPeL43, which was reported to be a potential marker of NPC (28).
RRAS was selected in our further study because it is down-regulated in NPC cell lines and tissues. The role of RRAS is controversial in different types of tumors. RRAS is overexpressed in a subset of gastric cancers by DNA demethylation and promotes cell survival (29). RRAS also act as an oncogene in the tumor growth of cervical epithelial cells and melanoma (5,7). However, Song et al. (6) reported that RRAS was reduced in breast cancer tissues and play a role as a tumor suppressor. Komatsu and Ruoslahti (9) also reported that RRAS suppressed intimal hyperplasia and tumor angiogenesis. The effects of RRAS may vary in different tumor types and stages. Up to now, the expression profiles and functions of RRAS in NPC has not been reported. In the present study, we confirmed that RRAS is down-regulated in NPC and might inhibit NPC proliferation and invasion. Our results indicate a tumor suppressor role of RRAS in NPC.
Different bioinformatics strategies have been reported in several studies to identify candidate genes associated with NPC. Hui et al. (30) performed a comprehensive analysis of methylation profiling and gene expression profiling to identify potentially critical differentially methylated genes in NPC. Wang et al. (31) analyzed the online dataset to find differentially expressed genes, and then GO, and KEGG analyses and the PPI network were applied to identify candidate genes related to EBV-associated NPC. We introduced a new strategy reported by Hu et al. (14) that has not been applied in NPC. Because the software GenRev is not available now, we adjusted our method to identify genes that interact with hub genes identified through pathway crosstalk and PPI analysis.
Our strategy provided a more targeted method that can eliminate the interference of much redundant information and has been proven to be feasible by biological experiments. It can be extended to analyze other pathways in our NPC pathway crosstalk module, and can also be applied to other diseases. However, there are still some limitations to our study. First, we only retrieved literature published on PubMed by October 10, 2018. Genes reported in some unpublished studies or those published after October 2018 were not included in our NPCgset. Therefore, our gene set needs to be constantly improved and updated. Second, although we have developed a series of criteria to select genes included in our study, false positives are still inevitable based on the uneven quality of the studies. Additionally, the results removed from our NPCgset according to our criteria might be proven to be associated with NPC in further studies. Third, in our pathway crosstalk analysis, the cell cycle and p53 signaling pathway were included but showed weaker crosstalk than other pathways involved in the two modules according to our algorithm; more studies are needed to identify potential connections between these pathways.
Conclusions
In our study, we extracted genes associated with NPC from publications in the PubMed database and constructed an NPCgset. GO, and KEGG pathway enrichment analysis was used to study the characteristics of genes in the NPCgset. Pathway Crosstalk Analysis showed that the pathways involved in the NPCgset could be summarized into two models: one module was related to the carcinogenesis process, and the other was correlated with the immune response. Furthermore, we constructed two PPI networks to search for new candidate genes potentially related to NPC. Finally, we found that RRAS was down-regulated in NPC, and the knockdown of RRAS promoted cell proliferation and invasion.
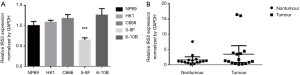
Table S1
| Gene set | Description | No. of genes | P value | FDR |
|---|---|---|---|---|
| GO:0001667 | Ameboidal-type cell migration | 46 | 0 | 0 |
| GO:0002521 | Leukocyte differentiation | 53 | 0 | 0 |
| GO:0006979 | Response to oxidative stress | 53 | 0 | 0 |
| GO:0007050 | Cell cycle arrest | 38 | 0 | 0 |
| GO:0007159 | Leukocyte cell-cell adhesion | 64 | 0 | 0 |
| GO:0007346 | Regulation of mitotic cell cycle | 67 | 0 | 0 |
| GO:0007568 | Aging | 52 | 0 | 0 |
| GO:0009314 | Response to radiation | 60 | 0 | 0 |
| GO:0009612 | Response to mechanical stimulus | 35 | 0 | 0 |
| GO:0009617 | Response to bacterium | 61 | 0 | 0 |
| GO:0010035 | Response to inorganic substance | 57 | 0 | 0 |
| GO:0022407 | Regulation of cell-cell adhesion | 53 | 0 | 0 |
| GO:0033002 | Muscle cell proliferation | 39 | 0 | 0 |
| GO:0033674 | Positive regulation of kinase activity | 57 | 0 | 0 |
| GO:0040017 | Positive regulation of locomotion | 60 | 0 | 0 |
| GO:0042493 | Response to drug | 58 | 0 | 0 |
| GO:0044770 | Cell cycle phase transition | 62 | 0 | 0 |
| GO:0045785 | Positive regulation of cell adhesion | 57 | 0 | 0 |
| GO:0045787 | Positive regulation of cell cycle | 48 | 0 | 0 |
| GO:0048514 | Blood vessel morphogenesis | 66 | 0 | 0 |
| GO:0048545 | Response to steroid hormone | 48 | 0 | 0 |
| GO:0048732 | Gland development | 59 | 0 | 0 |
| GO:0050673 | Epithelial cell proliferation | 56 | 0 | 0 |
| GO:0050900 | Leukocyte migration | 53 | 0 | 0 |
| GO:0051052 | Regulation of DNA metabolic process | 48 | 0 | 0 |
| GO:0051090 | Regulation of sequence-specific DNA binding transcription factor activity | 48 | 0 | 0 |
| GO:0051098 | Regulation of binding | 45 | 0 | 0 |
| GO:0051272 | Positive regulation of cellular component movement | 58 | 0 | 0 |
| GO:0070482 | Response to oxygen levels | 55 | 0 | 0 |
| GO:0070997 | Neuron death | 49 | 0 | 0 |
| GO:0071216 | Cellular response to biotic stimulus | 38 | 0 | 0 |
| GO:0071417 | Cellular response to organonitrogen compound | 55 | 0 | 0 |
| GO:0071900 | Regulation of protein serine/threonine kinase activity | 57 | 0 | 0 |
| GO:0090130 | Tissue migration | 41 | 0 | 0 |
| GO:0097191 | Extrinsic apoptotic signaling pathway | 42 | 0 | 0 |
| GO:0097193 | Intrinsic apoptotic signaling pathway | 41 | 0 | 0 |
| GO:2001233 | Regulation of apoptotic signaling pathway | 55 | 0 | 0 |
| GO:0031589 | Cell-substrate adhesion | 42 | 1.11E−16 | 2.23E−15 |
| GO:0007369 | Gastrulation | 32 | 2.22E−16 | 4.34E−15 |
| GO:0048144 | Fibroblast proliferation | 23 | 4.44E−16 | 8.46E−15 |
| GO:0018212 | Peptidyl-tyrosine modification | 45 | 6.66E−16 | 1.24E−14 |
| GO:0071559 | Response to transforming growth factor beta | 34 | 1.78E−15 | 3.22E−14 |
| GO:0001101 | Response to acid chemical | 41 | 2.44E−15 | 4.23E−14 |
| GO:0071214 | Cellular response to abiotic stimulus | 39 | 2.44E−15 | 4.23E−14 |
| GO:0002764 | Immune response-regulating signaling pathway | 50 | 3.33E−15 | 5.64E−14 |
| GO:0031349 | Positive regulation of defense response | 46 | 4.22E−15 | 6.99E−14 |
| GO:1901652 | Response to peptide | 48 | 7.55E−15 | 1.22E−13 |
| GO:0072331 | Signal transduction by p53 class mediator | 36 | 1.02E−14 | 1.62E−13 |
| GO:0001819 | Positive regulation of cytokine production | 44 | 1.19E−14 | 1.85E−13 |
Acknowledgments
We extend our sincere thanks to Professor Junfeng Xia (Institute of Health Sciences, Anhui University, Hefei) for his help in this study.
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.04.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Our research was approved by the Research Ethics Committee of Sun Yat-sen University Cancer Center (No. GZR2015-062), and all patients signed informed consent forms for publication of this manuscript and any accompanying images.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chua MLK, Wee JTS, Hui EP, et al. Nasopharyngeal carcinoma. Lancet 2016;387:1012-24. [Crossref] [PubMed]
- Zhang L, Huang Y, Hong S, et al. Gemcitabine plus cisplatin versus fluorouracil plus cisplatin in recurrent or metastatic nasopharyngeal carcinoma: a multicentre, randomised, open-label, phase 3 trial. Lancet 2016;388:1883-92. [Crossref] [PubMed]
- Wang M, Chen B, Chai L. Triptolide suppresses the proliferation and induces the apoptosis of nasopharyngeal carcinoma cells via the PI3K/Akt pathway. Oncol Lett 2019;17:1372-8. [PubMed]
- Liu WN, Yan M, Chan AM. A thirty-year quest for a role of R-Ras in cancer: from an oncogene to a multitasking GTPase. Cancer Lett 2017;403:59-65. [Crossref] [PubMed]
- Rincon-Arano H, Rosales R, Mora N, et al. R-Ras promotes tumor growth of cervical epithelial cells. Cancer 2003;97:575-85. [Crossref] [PubMed]
- Song J, Zheng B, Bu X, et al. Negative association of R-Ras activation and breast cancer development. Oncol Rep 2014;31:2776-2784. [Crossref] [PubMed]
- Gawecka JE, Griffiths GS, Ek-Rylander B, et al. R-Ras regulates migration through an interaction with filamin A in melanoma cells. PLoS One 2010;5:e11269. [Crossref] [PubMed]
- Wozniak MA, Kwong L, Chodniewicz D, et al. R-Ras controls membrane protrusion and cell migration through the spatial regulation of Rac and Rho. Mol Biol Cell 2005;16:84-96. [Crossref] [PubMed]
- Komatsu M, Ruoslahti E. R-Ras is a global regulator of vascular regeneration that suppresses intimal hyperplasia and tumor angiogenesis. Nature Medicine 2005;11:1346-50. [Crossref] [PubMed]
- Xu L, Gao Y, Chen Y, et al. Quantitative proteomics reveals that distant recurrence-associated protein R-Ras and Transgelin predict post-surgical survival in patients with Stage III colorectal cancer. Oncotarget 2016;7:43868-93. [PubMed]
- Wang MH, Sun R, Zhou XM, et al. Epithelial cell adhesion molecule overexpression regulates epithelial-mesenchymal transition, stemness and metastasis of nasopharyngeal carcinoma cells via the PTEN/AKT/mTOR pathway. Cell Death Dis 2018;9:2. [Crossref] [PubMed]
- Xu MJ, Johnson DE, Grandis JR. EGFR-targeted therapies in the post-genomic era. Cancer Metastasis Rev 2017;36:463-73. [Crossref] [PubMed]
- Zhang J, Jia L, Tsang CM, et al. EBV Infection and Glucose Metabolism in Nasopharyngeal Carcinoma. Adv Exp Med Biol 2017;1018:75-90. [Crossref] [PubMed]
- Hu Y, Pan Z, Hu Y, et al. Network and Pathway-Based Analyses of Genes Associated with Parkinson’s Disease. Mol Neurobiol 2017;54:4452-65. [Crossref] [PubMed]
- Jia P, Kao C, Kuo P, et al. A comprehensive network and pathway analysis of candidate genes in major depressive disorder. BMC Syst Biol 2011;5:S12. [Crossref] [PubMed]
- Shi L, Xiao R, Wang M, et al. MicroRNA-342-3p suppresses proliferation and invasion of nasopharyngeal carcinoma cells by directly targeting Cdc42. Oncol Rep 2018;40:2750-7. [PubMed]
- Stanczyk FZ, Mathews BW, Sherman ME. Relationships of sex steroid hormone levels in benign and cancerous breast tissue and blood: A critical appraisal of current science. Steroids 2015;99:91-102. [Crossref] [PubMed]
- Capper CP, Rae JM, Auchus RJ. The Metabolism, Analysis, and Targeting of Steroid Hormones in Breast and Prostate Cancer. Horm Cancer 2016;7:149-64. [Crossref] [PubMed]
- Dobruch J, Daneshmand S, Fisch M, et al. Gender and Bladder Cancer: A Collaborative Review of Etiology, Biology, and Outcomes. Eur Urol 2016;69:300-10. [Crossref] [PubMed]
- Dai W, Yin P, Chen P, et al. Study of urinary steroid hormone disorders: difference between hepatocellular carcinoma in early stage and cirrhosis. Anal Bioanal Chem 2014;406:4325-35. [Crossref] [PubMed]
- Hu Q, Lin X, Ding L, et al. ARHGAP42 promotes cell migration and invasion involving PI3K/Akt signaling pathway in nasopharyngeal carcinoma. Cancer Med 2018;7:3862-74. [Crossref] [PubMed]
- Lin YT, Wang HC, Hsu YC, et al. Capsaicin Induces Autophagy and Apoptosis in Human Nasopharyngeal Carcinoma Cells by Downregulating the PI3K/AKT/mTOR Pathway. Int J Mol Sci 2017; [Crossref] [PubMed]
- Ming H, Lan Y, He F, et al. Cytochrome b5 reductase 2 suppresses tumor formation in nasopharyngeal carcinoma by attenuating angiogenesis. Chin J Cancer 2015;34:459-67. [Crossref] [PubMed]
- Chang PY, Wu ZZ, Sun NK, et al. EBV-encoded LMP-1 sensitizes nasopharyngeal carcinoma cells to genotoxic drugs by down-regulating Cabin1 expression. J Cell Physiol 2014;229:309-22. [Crossref] [PubMed]
- You R, Hua Y, Liu Y, et al. Concurrent Chemoradiotherapy with or without Anti-EGFR-Targeted Treatment for Stage II-IVb Nasopharyngeal Carcinoma: Retrospective Analysis with a Large Cohort and Long Follow-up. Theranostics 2017;7:2314-24. [Crossref] [PubMed]
- Ooft ML, Braunius WW, Heus P, et al. Prognostic significance of the EGFR pathway in nasopharyngeal carcinoma: a systematic review and meta-analysis. Biomark Med 2015;9:997-1010. [Crossref] [PubMed]
- Hsu CH, Gao M, Chen CL, et al. Inhibitors of Epidermoid Growth Factor Receptor Suppress Cell Growth and Enhance Chemosensitivity of Nasopharyngeal Cancer Cells in vitro. Oncology 2005;68:538-47. [Crossref] [PubMed]
- Sim EU, Chan SL, Ng KL, et al. Narayanan K. Human Ribosomal Proteins RPeL27, RPeL43, and RPeL41 Are Upregulated in Nasopharyngeal Carcinoma Cell Lines. Dis Markers 2016;2016:5179594. [Crossref] [PubMed]
- Nishigaki M, Aoyagi K, Danjoh I, et al. Discovery of aberrant expression of R-RAS by cancer-linked DNA hypomethylation in gastric cancer using microarrays. Cancer Res 2005;65:2115-24. [Crossref] [PubMed]
- Hui L, Zhang J, Ding X, et al. Identification of potentially critical differentially methylated genes in nasopharyngeal carcinoma: A comprehensive analysis of methylation profiling and gene expression profiling. Oncol Lett 2017;14:7171-8. [PubMed]
- Wang J, Mei F, Gao X, et al. Identification of genes involved in Epstein-Barr virus-associated nasopharyngeal carcinoma. Oncol Lett 2016;12:2375-80. [Crossref] [PubMed]
(English Language Editor: Jeremy Dean Chapnick, AME Publishing Company)