
Acid-induced autophagy protects human gastric cancer cells from apoptosis by activating Erk1/2 pathway
Introduction
The most significant features of gastric cancer, the second most common cancer worldwide, are high mortality and poor prognosis (1,2). Currently, the available treatments include surgery, chemotherapy, radiation, and molecular targeted therapy. However, the prognoses of patients with gastric cancer remain relatively poor and the efficacy of chemotherapy and radiation treatments is limited by their side effects (3). Hence, a better understanding of gastric cancer progression is needed urgently.
Accumulating evidence reveals that the acidic tumor microenvironment plays a key role in tumor cell behavior (4). Rapid glycolysis together with poor tumor perfusion, result in lower pH of the extracellular tumor microenvironment (5). Previous research has shown that the pH of human tumor cells is for the most part, lower than that of normal tissues (6). As tumors grow rapidly, blood perfusion and oxygen supply are often insufficient, resulting in hypoxia and acidosis, which can be poisonous (7,8). In addition, extracellular acidification is suggested to be mutagenic and alters gene expression (9,10), leading to changes phenotype that are adapted for survival. Therefore, we propose that some adaptive mechanisms appear to promote cancer cell survival. However, the mechanism of how gastric cancer cells adapt to an acidic microenvironment remain unknown.
Determination of the mechanism of survival in low pH could provide information about tumor progression and facilitate the development of novel therapeutic methods. As the tumor microenvironment is acidic, hypoxic, and nutrient-deprived compared with its counterparts, these conditions can induce autophagy to promote tumor cell survival (11,12). We thus propose that autophagy can be stimulated by the acidic tumor microenvironment.
Autophagy, which digests damaged cytoplasmic proteins, macromolecules, and organelles (13), by packing them into double-membrane vesicles and transporting them to lysosomes for degradation (14), is a catabolic process through which cellular metabolism is maintained. According to previous studies, autophagy is a self-sufficient program that improves tumor cell viability by inhibiting apoptosis under adverse microenvironmental conditions (15,16). However, some studies indicate that autophagy induced in cancer cells inhibits the proliferation and growth of tumors (17). It is thus indispensable to determine the precise role of autophagy in gastric tumor cells subjected to an acidic microenvironment.
Activation of extracellular signal-regulated kinases Erk1/2 is involved in the induction of autophagy responses to stimuli including hypoxia (18) and curcumin (19). The Erk1/2 signaling pathway plays a crucial role in many cellular events including apoptosis and autophagy. In addition, Erk1/2 is associated with a variety of biological responses such as proliferation, migration, apoptosis, and autophagy (20). However, the role of the ERK1/2 pathway in low pH-induced autophagy is not known. Therefore, this study aimed to determine the role of Erk1/2 in acidic microenvironment-induced autophagy and the effect of autophagy on cytotoxicity mediated by low pH.
Methods
Cell culture
AGS cells were acquired from Cell Research Co. and were maintained in F12K (Cell Research, China, 500 mL) medium supplemented with 10% FBS (Gibco, New York, USA, 500 mL), 100 units/mL penicillin, and 100 µg/mL streptomycin (HyClone Laboratories, USA, 100 mL). The cells were maintained at 37 °C with 5% CO2. The cells were further exposed to 25 mmol/L each of PIPES (Thermo Fisher Scientific, China, 100 mL) and HEPES (Thermo Fisher Scientific, China, 100 mL) adjusted to pH 7.4 and 6.5 (21), and were monitored by microscopy to ensure that they maintained their original morphology. The pH of the medium in each group was measured before every experiment to prevent pH changes and ensure that the cells were exposed to stable experimental conditions. The cells were divided into two groups, which were incubated at pH 6.5 or 7.4.
Real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the cells using Trizol reagent (TaKaRa, Tokyo, Japan). One microgram of RNA per sample was reverse transcribed with Prime Script™ RT Reagent (TaKaRa, Tokyo, Japan) according to the manufacturers’ instructions. Reverse transcription was performed at 37 °C for 15 min, 84 °C for 5 s, and the samples were then stored at 4 °C.
mRNA expression was quantified using SYBR Green chemistry, as described previously (22). Expression of autophagy-related (including LC3, Beclin1, P62) and apoptosis-related (including Bax, Bcl2, caspase3) genes was normalized using β-actin. Upregulation or downregulation of mRNA expression was quantified by the cycle threshold (CT) method, and melting curve analysis was performed at the end of every RT-qPCR run. SPSS version 19.0 was used to calculate the statistical differences in the mRNA expression levels of various genes (23).
Western blot analysis
After cells were treated with different conditions, they were lysed in a lysis buffer including protease inhibitors. Protein concentrations were measured using a BCA kit. Twenty micrograms of protein per sample was separated on a 12% SDS gel, followed by semi-dry western blotting and transfer onto polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with 5% BSA in TBST [50 mM Tris–HCl (pH 7.5) and 150 mM NaCl containing 0.05% Tween 20] (24) for 1 h, and incubated overnight at 4 °C with primary antibodies against β-actin (1:1,000, rabbit, HuaBio), LC3B (1:1,000, rabbit, Bioworld), Beclin1 (1:1,000, mouse, Bioworld), SQSTM1/P62 (1:1,000, rabbit, CST), Bcl2 (1:1,000, rabbit, CST), cleaved caspase3 (1:1,000, rabbit, CST), Bax (1:1,000, rabbit, Bioworld), Erk1/2 (1:1,000, rabbit, CST), and p-Erk1/2 (1:1,000, rabbit, CST). Subsequently, the membranes were incubated with HRP-conjugated secondary antibodies (1:10,000 dilution, anti-rabbit or anti-mouse, CST) for 1 h at room temperature. Bands on the immunoblots were visualized with Millipore Immobilon ECL (Millipore, USA) and detected on Bio-Rad ChemiDoc™ XRS+.
Detection of mRFP-GFP-LC3
AGS cells treated at different pH (6.5 and 7.4) were seeded on coverslips and grown to reach 50% confluence. They were then infected with mRFP-GFP-LC3 adenoviruses (HanBio, shanghai) according to the manufacturer’s guidelines. The medium was replaced with different pH medium after 24 h. Cells were then fixed with 4% paraformaldehyde. The fixed cells were treated with 5% BSA for 30 min and stained with DAPI (Solarbio, Tongzhou Dist. Beijing) for 6 min. The coverslips were then observed by laser confocal microscopy (×189 magnification), as described previously (25). We counted the number of mRFP-GFP-LC3 puncta in three independent visual fields and analyzed the results using Image J software.
Transmission electron microscopy (TEM)
After the cells were seeded in media with different pH values (6.5 and 7.4) for 24 h, they were fixed overnight at 4 °C in 2.5% glutaraldehyde in 0.1 M phosphate buffer followed by post-fixation in 1% osmium tetroxide. After dehydration in a graded series of acetone, the cells were embedded in resin embedding medium (26). Finally, ultrathin sections were observed and imaged using a transmission electron microscope.
CCK8 assay for cell viability
Cell viability was evaluated by a cell counting kit 8 (CCK) assay (27). Cells exposed to media with different pH (6.5 and 7.4) were seeded in 96-well culture plates at a density of 4×103 cells/mL and incubated at 37 °C for approximately 12 h. The medium was then replaced with 100 µL of fresh medium containing 10% CCK8 (WST-8, Yiyuanbiotech). Cells were subsequently incubated for 0.5–3 h at 37 °C with 5% CO2, and absorbance was determined at 450 nm using a microplate spectrophotometer. Cell viability was calculated by the following formula: Cell viability (%) = [A450 (sample) − A450 (blank)/A450 (control) − A450 (blank)]×100%.
Cell migration and invasion assay
Cell invasion assays were performed using Transwell chambers (Corning Costar, USA). Briefly, cells in the logarithmic phase of growth were starved in serum-free F12K medium for 24 h, after which they were digested using 0.25% EDTA-trypsin. Cells were then suspended in serum-free F12K medium at a density of 5×105/mL. Approximately 200 µL of cell suspension was added to the upper chamber of the Transwell insert whose filter was coated with 80 µg of Matrigel (BD Biosciences, USA), and F12K medium containing 10% FBS was added to the lower chamber as a chemoattractant. Next, the cells were cultured for 24 h at 37 °C with 5% CO2. After 24 h, the chambers were fixed with methanol for 20 min and then stained with crystal violet for the same time. The non-invading cells and the Matrigel were removed with cotton swabs (28) and then the chamber was placed under an inverted microscope to count the remaining cells. Migration assays were performed using the same procedure as the Transwell invasion assays but without Matrigel-coated Transwell chambers. All experiments were repeated in triplicate.
Statistical analysis
All the values are expressed as mean ± SEM. Significant differences among three groups were examined using ANOVA and Student’s test, all statistical analyses were performed with SPSS version 15.0. Differences were considered statistically significant at P<0.05.
Results
Acidic stress induces autophagy in AGS gastric cancer cells
Previous studies have shown that acidic microenvironments drive autophagy in breast and lung cancer cells (5,26). However, whether autophagy occurs in AGS gastric cancer cells under acidic pH remains unclear. Therefore, we exposed the cells to pH 6.5 and 7.4 to determine whether low-pH enhances the expression of autophagy genes. We performed real-time quantitative RT-PCR analysis to determine the expression of LC3, Beclin1, and P62. The results showed that low mRNA expression levels of LC3 and Beclin1 in AGS cells incubated at pH 7.4; however, treatment with low pH resulted in significant increases in the mRNA expression levels of the above proteins compared to that in control cells. In addition, P62 mRNA expression levels were greatly decreased after treatment at low pH compared with those in control cells (P<0.05, Figure 1A). Moreover, we performed western blot analysis to determine the expression of essential protein markers of autophagy such as LC3, Beclin1, and P62. Increased LC3II expression commonly serves as an indicator of elevated autophagy levels in cell models. As shown in Figure 1B,C, the LC3II/LC3I ratio was significantly increased in the low pH-treated group. Consistent with this result, the level of Beclin1 was also increased obviously, whereas the autophagy substrate P62 was significantly reduced compared with that in the control group (P<0.05). Morphological analysis by TEM showed typical double-membrane vesicles in the AGS cells treated with the medium at pH 6.5, whereas there were very few vesicles in cells at pH 7.4 (Figure 1D). In summary, these results suggested that AGS cells treated with low pH displayed enhanced autophagy compared to cells under neutral conditions.
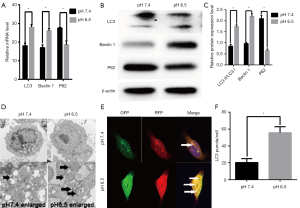
To confirm whether low pH treatment induces an autophagic flux, we transfected/AGS cells with mRFP-GFP-LC3B adenoviruses and monitored the mRFP-GFP-LC3 fluorescence using a fluorescence microscope. An emerging system involving the tandem fusion of LC3 to acid-resistant mCherry and acid-sensitive GFP has been applied for the analysis of autophagosome-mediated dynamic changes in proteins and protein degradation to monitor autophagic flux, as GFP fluorescence is quenched in low pH compartments. A dynamic switch from yellow to red fluorescence reflects a functional autophagic flux. In this system, LC3 is fused to both GFP and RFP to form an RFP-GFP-LC3 vector. As shown in Figure 1E,F cells treated with low-pH condition for 24 h displayed markedly increased numbers of red puncta stably expressing mRFP-GFP-LC3B compared to the control cells (P<0.05), suggesting that acidic stress increases autophagic flux. Taken together, our data suggest that low pH has dynamic effects on the autophagic flux.
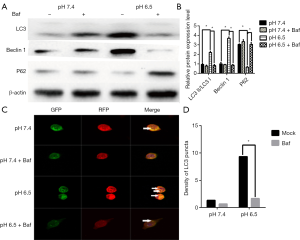
In addition, when cells were treated with the autophagy inhibitor Baf (10 mM, Sigma) at pH 6.5, we found that P62 expression was significantly increased whereas LC3 showed reduced (P<0.05, Figure 2A,B). When detecting the mRFP-GFP-LC3 fluorescence at pH 7.4 in the presence of Baf, we observed that the yellow fluorescent puncta almost did not switch to red puncta after 24 h. However, under pH 6.5 in the presence of Baf, the yellow puncta switched to red fluorescence (Figure 2C,D). These findings suggest that acidic stress can increases the autophagic flux in AGS gastric cancer cells.
Erk1/2 pathway participates in acid-induced autophagy in AGS gastric cancer cells
Previous results showed that activation of the MAPK Erk1/2 could lead to autophagy (29,30). Therefore, we determined the contribution of this pathway to acidic stress-induced autophagy. To detect whether acidic conditions induce autophagy through the rk1/2 pathway in AGS cells, we analyzed Erk1/2 and p-Erk1/2 by western blotting. We noted that when cells were exposed to an acidic microenvironment, p-Erk1/2 levels were increased compared to those in cells at pH 7.4 (Figure 3A,B).
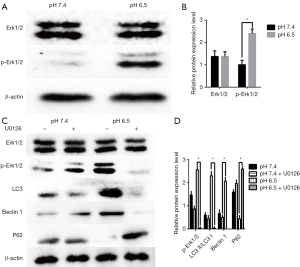
To investigate the relationships among acidic stress, the Erk1/2 pathway, and autophagy, we examined the effect of the MEK1/2 inhibitor, U0126. As shown in Figure 3C,D, U0126 obviously inhibited the expression of p-Erk1/2, as well as that of autophagy related proteins such as LC3 and Beclin1, whereas P62 expression was increased. Thus, inhibition of Erk1/2 resulted in decreased autophagy. These results indicate that acidic stress is mediated through Erk1/2 production and that the acidic microenvironment activates autophagy via the Erk1/2 pathway.
Acidic stress inhibits apoptotic cell death by enhancing autophagy induction in AGS cells
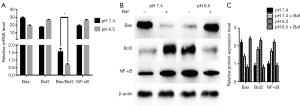
Apoptosis occurs via extrinsic, intrinsic, and ER stress-mediated processes (31,32). To determine whether apoptosis is mediated by autophagy induction in low pH-treated AGS cells, we first measured the expression of apoptosis-related mRNAs (such as Bax and Bcl2) in cells by RT-qPCR analyses. As shown in Figure 4A, acid-induced stress decreased Bax mRNA expression levels and the Bax/Bcl2 ratio, along with increased Bcl2 expression. As shown in Figure 4B,C, AGS cells treated with low-pH displayed significant decreases in Bax expression compared with cells at pH 7.4 (P<0.05). Besides, Bax expression decreased after culture in pH 7.4 with Baf treatment while increased in pH 6.5 with Baf. The above-mentioned apoptosis-related protein is pro-apoptotic protein and these results suggest that acidic stress inhibits apoptosis progression. In addition, we analyzed the expression of Bcl2 and NF-κB, which are anti-apoptotic proteins. As expected, we noted a clear increase in the levels of these proteins in the low pH-treated group. However, cells at neutral pH exhibited relatively weak Bcl2 and NF-κB expression (P<0.05). In order to explain the effect of the autophagy obstruction on the apoptosis progression, we evaluated the anti-apoptotic proteins with Baf, the Western blot results supplied that anti-apoptotic proteins in pH7.4 group with Baf were increased compared to the group without Baf. The expression of these proteins in the pH 6.5 group with Baf was opposed to the result of neutral pH-treated group, which showed decreased than the Baf-negative one. Collectively, these results suggest that acidic stress inhibits apoptosis.
Enhanced growth, invasion, and metastatic potential of AGS cells after acidic priming
Autophagy can be a pro-survival mechanism under different conditions (33). Simultaneously, previous studies have shown that acidic microenvironments can also drive tumor growth, invasion, and autophagy (34). To determine whether the acidic microenvironment promotes AGS cell growth and proliferation in vitro, we treated the cells with different pH conditions (pH 6.5 and 7.4). The CCK8 assay was conducted to measure cell viability. As shown in Figure 5A, low pH treatment greatly facilitated AGS cell growth whereas cells at pH 7.4 displayed barely detectable signs of toxicity, suggesting that this treatment had a slight cytostatic effect. Additionally, cell viability was lower in pH 6.5 than in pH 7.4 upon Baf treatment (P<0.05). This finding led us to conclude that acidic stress can effectively promote AGS cell viability.
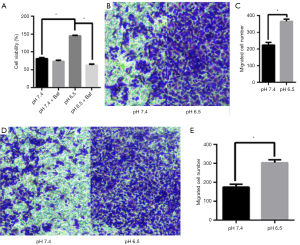
To evaluate the effect of acidic stress on AGS cell migration and invasion in an acidic microenvironment, we performed transwell assays. We found that low pH induced dramatic increases in both cell migration and invasion through Matrigel to the lower chamber in the Transwell assay compared with those in the control group (P<0.05, Figure 5B,C,D,E). These results indicate that acidic stress plays an important role in promoting the migration and invasion of AGS cells.
Discussion
Previous studies have shown that tumor cells exposed to acidic conditions undergo metabolic changes and that upregulation of glycolysis in these cells results in the extracellular accumulation of metabolic acids (35). Acidic pH has been shown to promote local invasive growth and metastasis. In addition, autophagy can be induced by extracellular acidosis and appears to serve as an adaptation enabling cells to survive in acidic microenvironments (36). Autophagy is a cellular catabolic process that induces the lysosomal degradation and recycling of dysfunctional proteins or organelles, which are considered vital processes to the survival of cancer cells exposed to acidic stress. We have determined that autophagy plays a complex role in tumor progression (14). In this study, we determined that autophagy acts as a protective mechanism by inhibiting apoptosis in gastric cancer cells in vitro. Moreover, our results indicate that acidic stress induces autophagy and that autophagy weakens the susceptibility of gastric cancer cells to acid-induced apoptosis. Furthermore, the Erk1/2 pathway was found to be involved in autophagy under an acidic microenvironment. Our experimental data demonstrate that AGS gastric cancer cells survive in an acidic microenvironment by activating autophagy through the Erk1/2 pathway.
One of the characteristics of tumors is that cancer cells can escape apoptosis and show increased growth (37). Autophagy can support this characteristic by playing a tumor survival role under metabolic stress such as an acidic microenvironment (26). Thus, there may be a link between the tumor acidic microenvironment and autophagy. In our study, we discussed the changes in gastric cancer cells under acidic stress. Finally, our initial hypothesis that acidic stress may induce autophagy in gastric cancer cells was verified by determining the protein expression of LC3 II, Beclin1, and P62 by western blotting. We found that the autophagy positive-correlated proteins, LC3II and Beclin1, were elevated under acidic stress whereas the autophagy negative-correlated protein P62 was inhibited. In addition, we found that in acidic microenvironment, LC3 puncta were increased compared with that in the control group after infection with GFP-mRFP-LC3 adenoviruses. We also observed double-membrane vesicles upon acidic treatment through TEM. These results are consistent with those of previous studies suggesting that acidic stress could favor autophagy (34).
Previous studies have shown that the MAPK (JNK1/2, Erk1/2, and p38) pathway is involved in the regulation of cell survival and cell death (38). The Erk1/2 pathway plays an important role in the invasion or metastasis of many cancers, such as oral cancer, liver cancer and lung tumor (39). It also has been reported that the Erk1/2 pathway can lead to autophagy activation in lung cancer cells and liver cancer cells (40,41). Activation of Erk1/2 is related to the accumulation of autophagosomes (42). In the present study, the expression of p-Erk1/2 was significantly increased upon low pH treatment. The Erk1/2 pathway inhibitor U0126 could reverse the autophagy activation in AGS cells subjected to an acidic microenvironment. These results explain that acidic stress promotes autophagy in gastric cancer cells via the Erk1/2 pathway. Baf, which interrupts autophagolysosome formation, is widely used as an autophagy inhibitor. Our study demonstrated that the effect of Baf in AGS cells treated with low pH was similar to that of the Erk1/2 pathway inhibitor U0126, which confirmed the positive effect of acidic stress on autophagy in gastric cancer cells. To sum up, the anti-proliferation and anti-migration effects of acidic stress are likely based on the activation of the Erk1/2 pathway in AGS cells, as shown in our study. Taken together, we provide evidence that an acidic microenvironment can promote autophagy by activating the Erk1/2 pathway.
Autophagy has dual roles in promoting or inhibiting cell death, depending on the cellular circumstance (43). In some cell types, autophagy is an indispensable mechanism by which apoptosis is suppressed (5,44). Our data suggest that autophagy induced by low pH treatment is related to the reduction of apoptosis in AGS cells. Thus, the increased level of autophagy resulting from acidic stress could result in decreased apoptotic cell death. To test this hypothesis, cells were treated with Baf, which had a suppressive effect on autophagy, thus enhancing apoptosis and cell viability in acid-treated gastric cancer cells. These results suggest that autophagy has a protective effect in AGS cells. In conclusion, when cells face an adverse external environment such as acidic stress, autophagy appears to act as a cell savior to facilitate gastric cancer cell survival through suppression of apoptosis.
Cancer cells can develop different migration patterns under diverse microenvironments (39). Previous studies have shown that extracellular acidosis may increase the migration and invasion rate of tumor cells (45). In this present study, we found that AGS gastric cancer cells showed increased migration and acidosis-induced invasion. The CCK-8 assay suggested that inhibition of autophagy suppressed the proliferation of AGS cells, suggesting that this type of autophagy is protective. In conclusion, treatment with low pH resulted in enhanced cell growth, migration, and invasion. Therefore, acidosis-induced autophagy serves as a pro-survival signal in AGS cells.
The relationship between autophagy and tumor development is still not understood well. In our study, we found that autophagy may promote apoptosis under an acidic tumor microenvironment. Therefore, reversing the acidic microenvironment may be a new strategy for treating gastric cancer, but still needs further testing in the clinic. There are also many shortcomings in our study. First, we only chose the AGS gastric cancer cells in our study and additional cell lines are needed in future studies. In addition, these results need to be confirmed in vivo using animal models. Taken together, our results demonstrate that acidic stress promotes AGS cell growth by promoting autophagy via the Erk1/2 pathway. However, the exact mechanisms by which low-pH microenvironments promote autophagy to increase AGS cell growth still need more research.
Acknowledgments
Funding: The authors gratefully acknowledge the financial support from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.07.42). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Hartgrink HH, Jansen EPM, van Grieken NCT, et al. Gastric cancer. Lancet 2009;374:477-90. [Crossref] [PubMed]
- Ashraf N, Hoffe S, Kim R. Adjuvant Treatment for Gastric Cancer_ Chemotherapy Versus Radiation. Oncologist 2013;18:1013-21. [Crossref] [PubMed]
- Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013;501:346-54. [Crossref] [PubMed]
- Xie WY, Zhou XD, Li Q, et al. Acid-induced autophagy protects human lung cancer cells from apoptosis by activating ER stress. Exp Cell Res 2015;339:270-9. [Crossref] [PubMed]
- Calorini L. Francesca Bianchini. Extracellular acidity as favouring factor of tumor progression and metastatic dissemination. Exp Oncol 2012;34:79-84. [PubMed]
- Span PN, Bussink J. Biology of hypoxia. Semin Nucl Med 2015;45:101-9. [Crossref] [PubMed]
- Schornack PA, Gillies RJ. Contributions of Cell Metabolism and H+ Diffusion to the Acidic pH of Tumors. Neoplasia 2003;5:135-45. [Crossref] [PubMed]
- Chen JL, Lucas JE, Schroeder T, et al. The genomic analysis of lactic acidosis and acidosis response in human cancers. PLoS Genet 2008;4:e1000293. [Crossref] [PubMed]
- Gerweck LE, Seetharaman K. Cellular pH Gradient in Tumor versus Normal Tissue: Potential Exploitation for the Treatment of Cancer. Cancer Res 1996;56:1194-8. [PubMed]
- Yang X, Yu DD, Yan F, et al. The role of autophagy induced by tumor microenvironment in different cells and stages of cancer. Cell Biosci 2015;5:14. [Crossref] [PubMed]
- Rankin EB, Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ 2008;15:678-85. [Crossref] [PubMed]
- Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self-digestion. Nature 2008;451:1069-75. [Crossref] [PubMed]
- Hurley JH, Young LN. Mechanisms of Autophagy Initiation. Annu Rev Biochem 2017;86:225-44. [Crossref] [PubMed]
- Schmukler E, Grinboim E, Schokoroy S, et al. Ras inhibition enhances autophagy, which partially protects cells from death. Oncotarget 2013;4:145-55. [Crossref] [PubMed]
- Liu D, Yang Y, Liu Q, et al. Inhibition of autophagy by 3-MA potentiates cisplatin-induced apoptosis in esophageal squamous cell carcinoma cells. Med Oncol 2011;28:105-11. [Crossref] [PubMed]
- Li C, Wang Y, Wang C, et al. Anticancer activities of harmine by inducing a pro-death autophagy and apoptosis in human gastric cancer cells. Phytomedicine 2017;28:10-8. [Crossref] [PubMed]
- Wu J, Niu J, Li X, et al. Hypoxia induces autophagy of bone marrow-derived mesenchymal stem cells via activation of ERK1/2. Cell Physiol Biochem 2014;33:1467-74. [Crossref] [PubMed]
- Li X, Feng K, Li J, et al. Curcumin Inhibits Apoptosis of Chondrocytes through Activation ERK1/2 Signaling Pathways Induced Autophagy. Nutrients 2017;9: [Crossref] [PubMed]
- Pallichankandy S, Rahman A, Thayyullathil F, et al. ROS-dependent activation of autophagy is a critical mechanism for the induction of anti-glioma effect of sanguinarine. Free Radic Biol Med 2015;89:708-20. [Crossref] [PubMed]
- Riemann A, Schneider B, Gundel D, et al. Acidic priming enhances metastatic potential of cancer cells. Pflugers Arch 2014;466:2127-38. [Crossref] [PubMed]
- Karger S, Weidinger C, Krause K, et al. FOXO3a: a novel player in thyroid carcinogenesis? Endocr Relat Cancer 2009;16:189-99. [Crossref] [PubMed]
- Yu M, Gou WF, Zhao S, et al. Beclin 1 expression is an independent prognostic factor for gastric carcinomas. Tumour Biol 2013;34:1071-83. [Crossref] [PubMed]
- Zhang Z, Lai Q, Li Y, et al. Acidic pH environment induces autophagy in osteoblasts. Sci Rep 2017;7:46161. [Crossref] [PubMed]
- Maejima Y, Kyoi S, Zhai P, et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med 2013;19:1478-88. [Crossref] [PubMed]
- Wojtkowiak JW, Rothberg JM, Kumar V, et al. Chronic autophagy is a cellular adaptation to tumor acidic pH microenvironments. Cancer Res 2012;72:3938-47. [Crossref] [PubMed]
- Jing X, Xu Y, Cheng W, et al. Tanshinone I induces apoptosis and pro-survival autophagy in gastric cancers. Cancer Chemother Pharmacol 2016;77:1171-81. [Crossref] [PubMed]
- Zhao X, Li DC, Zhu XG, et al. B7-H3 overexpression in pancreatic cancer promotes tumor progression. Int J Mol Med 2013;31:283-91. [Crossref] [PubMed]
- Sivaprasad U, Basu A. Inhibition of ERK attenuates autophagy and potentiates tumour necrosis factor-alpha-induced cell death in MCF-7 cells. J Cell Mol Med 2008;12:1265-71. [Crossref] [PubMed]
- Su Z, Wang T, Zhu H, et al. HMGB1 modulates Lewis cell autophagy and promotes cell survival via RAGE-HMGB1-Erk1/2 positive feedback during nutrient depletion. Immunobiology 2015;220:539-44. [Crossref] [PubMed]
- Liu LQ, Fan ZQ, Tang YF, et al. The resveratrol attenuates ethanol-induced hepatocyte apoptosis via inhibiting ER-related caspase-12 activation and PDE activity in vitro. Alcohol Clin Exp Res 2014;38:683-93. [Crossref] [PubMed]
- Yuzefovych LV, Musiyenko SI, Wilson GL, et al. Mitochondrial DNA damage and dysfunction, and oxidative stress are associated with endoplasmic reticulum stress, protein degradation and apoptosis in high fat diet-induced insulin resistance mice. PLoS One 2013;8:e54059. [Crossref] [PubMed]
- Degenhardt K, Mathew R, Beaudoin B, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006;10:51-64. [Crossref] [PubMed]
- Marino ML, Pellegrini P, Di Lernia G, et al. Autophagy is a protective mechanism for human melanoma cells under acidic stress. J Biol Chem 2012;287:30664-76. [Crossref] [PubMed]
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer 2004;4:891-9. [Crossref] [PubMed]
- Estrella V, Chen T, Lloyd M, et al. Acidity generated by the tumor microenvironment drives local invasion. Cancer Res 2013;73:1524-35. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Lai KC, Huang AC, Hsu SC, et al. Benzyl isothiocyanate (BITC) inhibits migration and invasion of human colon cancer HT29 cells by inhibiting matrix metalloproteinase-2/-9 and urokinase plasminogen (uPA) through PKC and MAPK signaling pathway. J Agric Food Chem 2010;58:2935-42. [Crossref] [PubMed]
- Li PY, Lv J, Qi WW, et al. Tspan9 inhibits the proliferation, migration and invasion of human gastric cancer SGC7901 cells via the ERK1/2 pathway. Oncol Rep 2016;36:448-54. [Crossref] [PubMed]
- Yang L, Su T, Lv D, et al. ERK1/2 mediates lung adenocarcinoma cell proliferation and autophagy induced by apelin-13. Acta Biochim Biophys Sin (Shanghai) 2014;46:100-11. [Crossref] [PubMed]
- Zhang X, Yang H, Yue S, et al. The mTOR inhibition in concurrence with ERK1/2 activation is involved in excessive autophagy induced by glycyrrhizin in hepatocellular carcinoma. Cancer Med 2017;6:1941-51. [Crossref] [PubMed]
- Kun Z, Hanqing G, Hailing T, et al. Gastrin enhances autophagy and promotes gastric carcinoma proliferation via inducing AMPKalpha. Oncol Res 2017;25:1399-407. [Crossref] [PubMed]
- Maiuri MC, Zalckvar E, Kimchi A, et al. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007;8:741-52. [Crossref] [PubMed]
- Kaminskyy VO, Piskunova T, Zborovskaya IB, et al. Suppression of basal autophagy reduces lung cancer cell proliferation and enhances caspase-dependent and -independent apoptosis by stimulating ROS formation. Autophagy 2012;8:1032-44. [Crossref] [PubMed]
- Riemann A, Schneider B, Gundel D, et al. Acidosis Promotes Metastasis Formation by Enhancing Tumor Cell Motility. Adv Exp Med Biol 2016;876:215-20. [Crossref] [PubMed]


