
c[RGDyk]-coated liposomes loaded with adriamycin and miR-21 mimics inhibit the growth of hepatoma cell line SMCC-7721 via up-regulating Bax and p53
Introduction
Liver cancer, one of the most common cancers, mostly results from hepatitis and cirrhosis. It has high malignancy, low cure rate and poor prognosis with very high mortality. The death rate ranks the second in tumor-related deaths. The occurrence and development of liver cancer seriously harm the health and the quality of life of human (1-3). The pathogenesis of liver cancer is very complicated, which involves many factors such as the regulation of the relevant signaling pathways. Not only that, it also includes cell proliferation, apoptosis, invasion and migration. Primary liver cancer is usually divided into three types: hepatocellular carcinoma, cholangiocarcinoma, and mixed cell carcinoma. It is one of the most lethal cancers today, of which hepatocellular carcinoma accounts for 85–90% (4). Liver cancer is currently treated by surgery, interventional therapy, radiotherapy and chemotherapy, and targeted therapy. However, the current drug treatments are not satisfactory for liver cancer (5,6). Targeted nanodrug delivery system uses new drug loading system to increase the selectivity to the target lesions, thereby increasing the drug concentration in target tissues (7,8). Drug delivery system targeting the tumor cells would deliver more drug to the target cells, reduce the damage to normal cells, and increase the efficacy of drug therapy (9-11).
miRNA, a class of non-coding single strand small molecule RNA, is widely found in multicellular organisms. miRNA has a length of about 20 nucleotides, accounting for about 1% of the total number of human genes, forming regulatory networks that control the life of organisms (12-14).
The miR-21 gene is located on plus strand of chromosome 17q23.2 and is in the tenth intron of the TMEM49 gene. If abnormally expressed, it can promote the growth of hepatoma cells, indicating that miR-21 can affect the occurrence of liver cancer (15). At the same time, miR-21 can regulate target genes to participate in tumor development, such as tumor proliferation, apoptosis, invasion and metastasis. At the present stage, liver cancers are mainly treated with surgery, interventional therapy, radiotherapy and chemotherapy, as well as targeted therapy. However, the overall outcomes have been unsatisfactory and prognosis is still poor (16).
The targeted drug delivery using nano-sized liposomes is a new drug delivery approach. Studies have shown that this approach increases the targeting ability of the drugs to be delivered, thus the drug concentration in the targeted lesion or targeted cells, yielding improved therapeutic effect (6,17). In this study, we aimed to investigate the effect of miR-21mimics and liposome-loaded drug (nanodrug) on liver cancer using cell line SMCC-7721. The treated cells were investigated for cell cycle arrest and apoptosis, as well as the expression of apoptosis-related proteins. The findings would provide clues for targeted treatment of liver cancer.
Methods
Cell line
Liver cancer cell line SMCC-7721was purchased from the Cell Bank, Chinese Academy of Sciences, Shanghai, China and maintained in RPMI-1640 medium in 5% CO2 at 37 °C.
Reagents and instruments
Cell cycle staining kit (CCS012) and Annexin V-FITC/PI apoptosis kit (AP101-100-kit) were purchased from Multi-Sciences, Beijing, China. Trizon Reagent (CW0580S), ultrapure RNA extraction kit (CW0581M), HiFiScript first strand cDNA synthesis kit (CW2569M), miRNA purification and reverse transcription kit (CW0627S), ultraSYBR mixture (CW0957M) and BCA protein quantification kit (CW0014S) were products of CWBIO, Beijing, China. Ultrasensitive luminescence solution (RJ239676) was from Thermo Fisher, USA. Mouse monoclonal antibody against GAPDH (TA-08, 1:1,000), goat anti rabbit IgG (ZB-2301, 1:500) and goat anti mouse IgG (ZB-2305, 1:500) were purchased from Zsbio, Beijing, China. Mouse monoclonal antibody against P53 (cat no. 2524) was obtained from Cell Signaling, USA. Rabbit monoclonal antibody against Bax (ab32503) and polyclonal antibody against Bcl-2 were from Abcam, UK and Bioss, USA, respectively.
Flow cytometer (NovoCyteTM) was from Eisen Biologicals, Hangzhou, China. Fluorescent PCR instrument (CFX Connect™) and ultrasensitive chemiluminescence imaging system (Chemi DocTM XRS+) were purchased from Bio-Rad, Shanghai, China.
Drug treatments and transfection
c[RGDyk]-coated liposome loaded with adriamycin (nanodrug) was prepared as described previously (18) and miR-21 mimics was synthesized at Sangon Biotech, Shanghai, China. Cells were treated with 0 to 10 µL nanodrug/well at five concentrations (2.5 to 20 mol/L) and harvested 24, 48 and 72 h after for viability tests. For transfection, cells were grown to a confluency of 90% in RPMI 1640 medium and transfected with 125 µL Opti-MEM and 5 µL lipofectamine 3000 containing miRNA mimics or empty vector according to manufacturer’s protocols. Two h later, the transfected cells were treated with nanodrug for 72 h. Un-treated cells were used as control.
Flow cytometry
For cell cycle analysis, cells were suspended in PBS and added with DNA staining solution. After incubated at room temperature for 10 min, the cells were analyzed using a flow cytometer. For apoptosis detection, SMMC-7721 cells were harvested and washed in pre-chilled PBS twice, suspended in apoptosis positive control solution and incubated on ice for 30 min according to the manufacturer’s instructions. The cells were labeled with Annexin V and V-FITC following the manufacturer’s instructions and loaded onto flow cytometer to assess the apoptotic cells. The quantitation of apoptotic cells was calculated by CellQuest software (Becton Dickinson).
RT-PCR for gene expression
Total RNA was isolated from SMMC-7721 cells using ultrapure RNA extraction kit according to manufacturer’s recommendations. The RNA was quantified and reversely transcripted using HiFiScript cDNA synthesis kit based on manufacturer’s recommendations. The resulting cDNA was subjected to amplification using UltraSYBR Mixture (Applied Biosystems) in a total volume of 12 µL on CFX Connect™ PCR system. The cycling conditions were 50 °C for 2 min, 95 °C for 10 min followed by 40 cycles, each one consisting of 15 s at 95 °C and 1 min at 60 °C using the primers listed in Table 1. GAPDH was used as internal control. Samples were run in triplicate and the mean value was calculated for each case. The data were analyzed using the Applied Biosystems software RQ Manager v1.2.1. Relative expression was calculated by using comparative Ct method and obtaining the fold change value (2−ΔΔCt) according to previously described protocol (19).
Table 1
| Primer | Sequence |
|---|---|
| Mir-21 F | CGCCGTAGCTTATCAGACTG |
| Mir-21 R | CAGCCACAAAAGAGCACAAT |
| U6 F | CTCGCTTCGGCAGCACA |
| U6 R | AACGCTTCACGAATTTGCGT |
| BCL-2 F | GTGCCTGCTTTTAGGAGACCGA |
| BCL-2 R | GAGACCACACTGCCCTGTTGATC |
| P53 F | AGTGCTCGCTTAGTGCTCCCT |
| P53 R | GTGCATGTTTGTGCCTGTCCT |
| Bax F | AGACACTCGCTCAGCTTCTTG |
| Bax R | CTTTTGCTTCAGGGTTTCATC |
| GAPDH F | GAAGGTCGGAGTCAACGGAT |
| GAPDH R | CCTGGAAGATGGTGATGGG |
Western blot analysis
SMMC-7721 cells were added with 400 µL lysis buffer and protein lysates were harvested using RIPA buffer (50 mM Tris, pH 7.2; 150 mM NaCl; 1% Triton X-100; and 0.1% SDS) containing protease (1:100, Roche, USA) and phosphatase (1:100, Sigma-Aldrich, USA) inhibitors. The protein concentrations were determined using a BCA kit according to manufacturer’s instructions. Sixty µg proteins were separated on 12% SDS-PAGE and transferred to PVDF membranes. Protein expression levels were quantified using appropriate antibodies specific for each protein. The expression levels of these proteins were standardized to GAPDH. Primary antibodies were detected using goat anti-rabbit or goat anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5,000, Santa Cruz Biotechnology, USA). Immunoreactive bands were visualized and quantified by densitometry using chemiluminescence imaging system according to the manufacturer’s instructions.
Statistical analysis
All data were expressed as means ± standard error of the mean (SEM) obtained from at least three independent experiments. Statistical comparisons between experimental and control groups were assessed by using the Student’s t-test. P<0.05 was considered statistically significant.
Results
Effect of drug concentration and exposure time on cell viability
We first determined the effect of drug concentration on cell viability. As shown in Figure 1A, the viability of SMMC-7721 cells declined significantly when 2.5 µL nanodrug (at 1 mol/L) was added to the culture well. However, the viability did not reduce further as the amount added increased to 20 µL/well. Therefore, we choose to use 5 µL/well in subsequent study.
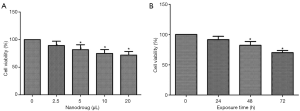
We then tested the impact of exposure time on the viability of SMMC-7721 cells. Measurements showed that within 72 h, the viability was not affected by the exposure time (Figure 1B). Therefore, in subsequent experiments, the cells were exposed to the drugs for 72 h.
Cell viability after transfection and drug treatment
We then analyzed the viability of SMMC-7721 cells after transfection and nanodrug treatments. Results showed that both miR-21 mimics and drug reduced the viability, but there was no synergism between them. The viabilities were similar between single and combined treatments (Figure 2). On other hand, empty vector did reduce the viability, but not as much as the expression vector (Figure 2).

Cell cycle
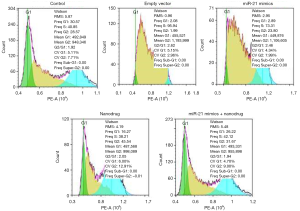
The flow cytometry results showed that there were more cells in S phase after nanodrug treatments. Compared with the negative control, there were more cells in S phase after the over-expression + nanodrug treatment, while the cells in the over-expression were more in G2 phase (Figure 3).
Apoptosis
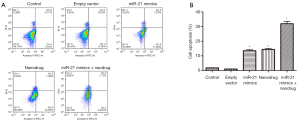
Apoptosis in untreated cells or cells treated with empty vector was low. miR-21 mimics + nanodrug treatment resulted significantly higher apoptotic rate as compared with other treatments, while the apoptotic rates were similar between single miR-21 mimics and nanodrug treatments (Figure 3).
Expression of genes
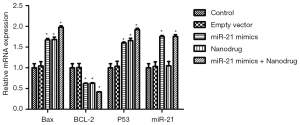
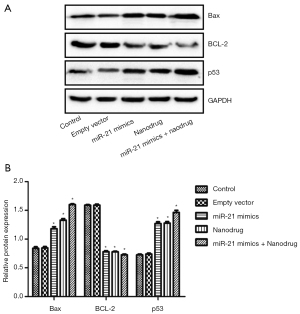
RT-PCR analysis showed that compared with the control, the expression of Bax and p53 in cells treated with nanodrug, miR-21 mimics + nanodrug and miR-21 mimics was significantly up-regulated, while that of Bcl-2 was significantly down-regulated at mRNA level (Figure 4). Similar changes were observed at protein levels for these genes (Figures 5,6). Meanwhile, miR-21 levels were increased after the cells were transfected with miR-21 mimics (Figure 5).
Discussion
Studies have shown that miRNAs are often abnormally expressed in cancer and play regulatory role on the expression of tumor-related genes and the occurrence, development and prognosis of tumor (20). miRNAs not only act as a proto oncogene, but also act as a tumor suppressors to regulate the related biological functions (21). For example, Gabriely et al. found that in glioma cells miR-21 participates in the regulation of invasion, migration. It regulates the expression of apoptosis-related genes, including TIMP3, BECK, and MMPs. MiR-21 downregulates the suppressor of MMPs, resulting activated MMPs to promote cell invasion and increase the malignancy of tumor (22). Data also show that miR-21 regulates Bcl-2 and other related genes in breast cancer, affecting the occurrence and development of tumors. In metastatic breast cancer cells, miR-21 inhibits the expression of PDCD4, TPM 1 and Maspin genes to impact tumor invasion and metastasis (23). Meng et al. found that the inhibition of miR-21 in hepatoma cells increases the expression of the PTEN gene, affecting the proliferation, migration and invasion of the tumor cells (15). MiRNA-21 is highly expressed in most tumors, such as gastric cancer (24), myeloma (25) and liver cancer (26).
In this study, we exposed hepatoma cells to nanodrug or transfected them with miR-21 mimics. The findings show that these treatments reduced the viability and increased the apoptosis of treated cells. At molecular level, Bax and p53 expression was upregulated and Bcl-2 expression was down-regulated. Bax, p53 and Bcl-2 are all apoptosis-related proteins. p53 directly regulates the expression of Bcl-2 and Bax is considered to be an apoptosis-promoting gene that induces apoptosis of tumor cells (11). Therefore, our findings suggest that miR-21 suppresses the growth of liver cancer cells via regulating the expression of apoptosis-related proteins and use of nanodrug and miR-21 may potentially offer new therapeutic options.
Apoptotic process is regulated by many genes in cells. Bcl-2 gene family is the most important family of apoptotic regulatory genes (27). It is closely related to apoptosis and is the most important gene to inhibit apoptosis of cancer cells. In addition to Bcl-2 gene, the Bcl-2 gene family also has Bax gene, which is an apoptotic promoter gene and plays an important role in the process of apoptosis. The ratio of Bax gene to Bcl-2 gene is an important factor determining apoptosis (28). Bax gene and Bcl-2 gene are the most representative anti-apoptotic and pro-apoptotic genes in Bcl-2 family, respectively. Bcl-2 is an important proto-oncogene encoding a protein with a relative molecular weight of 2500. Bax is one of the most widely studied apoptotic proteins in the Bcl-2 family. It can form heterodimer with Bcl-2 or homologous dimer itself. Bax is highly expressed in many apoptotic cells. Studies have confirmed that high expression of Bcl-2 inhibits the apoptosis induced by many factors. Increased expression of Bax induces cell apoptosis by inhibiting the activity of Bcl-2 (29). In addition, it is reported that the ratio of Bax to Bcl-2, rather than Bcl-2 alone, plays a decisive role in drug-induced apoptosis (30). More than 50% of human tumors are related to P53 mutations. P53 is the most closely related gene to human tumors. Different tumors have different mutation sites, showing the specificity of mutation (31). Mutational diversity exists even in different individuals of the same tumor. The point mutation rate of P53 gene in primary liver cancer was 25–60% (32).
Since this is an in vitro study and used only one cell line, these findings need to be further validated in vivo with more cell lines, particularly with non-tumorogenic hepatic cell line to demonstrate their specificity.
Acknowledgments
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.07.29). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were approved by the institutional ethics board, in compliance with the national and institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Glantzounis GK, Kyrochristos ID, Ziogas DE, et al. Novel translational therapeutic strategy by sequencing primary liver cancer genomes. Future Oncol 2017;13:1049-52. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Yang B, Petrick JL, Kelly SP, et al. Adiposity across the adult life course and incidence of primary liver cancer: The NIH-AARP cohort. Int J Cancer 2017;141:271-8. [Crossref] [PubMed]
- Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer 2006;6:674-87. [Crossref] [PubMed]
- Yegin EG, Oymaci E, Karatay E, et al. Progress in surgical and nonsurgical approaches for hepatocellular carcinoma treatment. Hepatobiliary Pancreat Dis Int 2016;15:234-56. [Crossref] [PubMed]
- Zeng S, Wu F, Li B, et al. Synthesis, characterization, and evaluation of a novel amphiphilic polymer RGD-PEG-Chol for target drug delivery system. ScientificWorldJournal 2014;2014:546176. [Crossref] [PubMed]
- Mirabello G, Lenders JJ, Sommerdijk NA. Bioinspired synthesis of magnetite nanoparticles. Chem Soc Rev 2016;45:5085-106. [Crossref] [PubMed]
- Ma X, Gong N, Zhong L, et al. Future of nanotherapeutics: Targeting the cellular sub-organelles. Biomaterials 2016;97:10-21. [Crossref] [PubMed]
- Sachdeva MS. Drug targeting systems for cancer chemotherapy. Expert Opin Investig Drugs 1998;7:1849-64. [Crossref] [PubMed]
- Katanasaka Y, Ishii T, Asai T, et al. Cancer antineovascular therapy with liposome drug delivery systems targeted to BiP/GRP78. Int J Cancer 2010;127:2685-98. [Crossref] [PubMed]
- Temiz P, Akkas G, Nese N, et al. Determination-of apoptosis and cell cycle modulators (p16, p21, p27, p53, BCL-2, Bax, BCL-xL, and cyclin D1) in thyroid follicular carcinoma, follicular adenoma, and adenomatous nodules via a tissue microarray method. Turk J Med Sci 2015;45:865-71. [Crossref] [PubMed]
- Siew WH, Tan KL, Babaei MA, et al. MicroRNAs and intellectual disability (ID) in Down syndrome, X-linked ID, and Fragile X syndrome. Front Cell Neurosci 2013;7:41. [Crossref] [PubMed]
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834-8. [Crossref] [PubMed]
- Hsu SH, Ghoshal K. MicroRNAs in Liver Health and Disease. Curr Pathobiol Rep 2013;1:53-62. [Crossref] [PubMed]
- Meng F, Henson R, Wehbe-Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007;133:647-58. [Crossref] [PubMed]
- Hu KQ. Advances in clinical application of cryoablation therapy for hepatocellular carcinoma and metastatic liver tumor. J Clin Gastroenterol 2014;48:830-6. [Crossref] [PubMed]
- Danhier F, Le Breton A, Preat V. RGD-based strategies to target alpha(v) beta(3) integrin in cancer therapy and diagnosis. Mol Pharm 2012;9:2961-73. [Crossref] [PubMed]
- Wei X, Zhou X, Tu Q, et al. Therapeutic effect of hepatocellular carcinoma-targeting liposome delivery system loaded with c[RGDyk] modified combretastatin A-4 and adriamycin: a pharmacodynamics study. Int J Clin Exp Pathol 2017;10:1417-22.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Aigner A. MicroRNAs (miRNAs) in cancer invasion and metastasis: therapeutic approaches based on metastasis-related miRNAs. J Mol Med (Berl) 2011;89:445-57. [Crossref] [PubMed]
- Manikandan J, Aarthi J, Kumar S, et al. Oncomirs: the potential role of non-coding micro RNAs in understanding cancer. Bioinformation 2008;2:330-4. [Crossref] [PubMed]
- Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol 2008;28:5369-80. [Crossref] [PubMed]
- Zhu S, Wu H, Wu F, et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 2008;18:350-9. [Crossref] [PubMed]
- Chan SH, Wu CW, Li AF, et al. miR-21 microRNA expression in human gastric carcinomas and its clinical association. Anticancer Res 2008;28:907-11. [PubMed]
- Pichiorri F, Suh SS, Ladetto M, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci U S A 2008;105:12885-90. [Crossref] [PubMed]
- Connolly E, Melegari M, Landgraf P, et al. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol 2008;173:856-64. [Crossref] [PubMed]
- Ashkenazi A, Fairbrother WJ, Leverson JD, et al. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors. Nat Rev Drug Discov 2017;16:273-84. [Crossref] [PubMed]
- Zhang Z, Liang Z, Li H, et al. Perfluorocarbon reduces cell damage from blast injury by inhibiting signal paths of NF-kappaB, MAPK and Bcl-2/Bax signaling pathway in A549 cells. PLoS One 2017;12:e0173884. [Crossref] [PubMed]
- Lin CH, Wu MR, Li CH, et al. Editor's Highlight: Periodic Exposure to Smartphone-Mimic Low-Luminance Blue Light Induces Retina Damage Through Bcl-2/BAX-Dependent Apoptosis. Toxicol Sci 2017;157:196-210. [Crossref] [PubMed]
- Wei L, Chen Q, Guo A, et al. Asiatic acid attenuates CCl4-induced liver fibrosis in rats by regulating the PI3K/AKT/mTOR and Bcl-2/Bax signaling pathways. Int Immunopharmacol 2018;60:1-8. [Crossref] [PubMed]
- Yue X, Zhao Y, Xu Y, et al. Mutant p53 in Cancer: Accumulation, Gain-of-Function, and Therapy. J Mol Biol 2017;429:1595-606. [Crossref] [PubMed]
- Wang S, Zhao Y, Aguilar A, et al. Targeting the MDM2-p53 Protein-Protein Interaction for New Cancer Therapy: Progress and Challenges. Cold Spring Harb Perspect Med 2017;7: [Crossref] [PubMed]


