
Knockdown of METTL14 inhibits the growth and invasion of cervical cancer
Introduction
Cervical cancer is the fourth most common malignancy of the female reproductive tract, with an estimated 530,000 new cases each year. Among the 530,000 cases of cervical cancer, there are approximately 270,000 deaths (1,2). In recent years, there has been an increase in the occurrence and mortality of cervical cancer. It is estimated that about 85% of cervical cancer deaths occur in underdeveloped or developing countries (1). As a result of technological advances in treatment strategies including surgery and radiotherapy, there has been an improvement in the 3-year local control rate of cervical cancer patients. However, the 5-year survival rate of patients in underdeveloped countries remains at <50% (1,3-5). Persistent infection with high risk human papillomavirus (HR-HPV) is the most important risk factor for cervical precancerous lesions and cervical cancer. It is reported that HPV DNA is identified in about 95% of malignant cervical lesions (6). Specifically, persistent overexpression of E6 and E7 oncogenes of HPV demonstrates a close relationship with the progression of cervical cancer (7). Although the sensitivity of HR-HPV detection has improved in recent years, nearly 15% of cervical cancer patients are HPV-negative that cannot be detected (8,9). In order to develop new therapeutic strategies for HPV-positive and HPV-negative cervical cancer, it is urgent to investigate the underlying mechanisms of cervical cancer and identify novel target genes.
Chemical modification of RNA is a common process in eukaryotes that impairs gene expression. One particular method of chemical modification is N6-methyladenosine (m6A) modification, which has been revealed to be involved in many biological functions in mammals, such as transcription splicing (10), DNA damage response (11), protein translation control (12) and cell fate determination (13,14). N6-methyladenosine is one of the most prevalent chemical markers for human mRNA. The heterodimer formed by Methyltransferase-like 3 (METTL3)/Methyltransferase-like 14 (METTL14) is the key protein for m6A modification. METTL3 contains the active methyltransferase domain catalyzing the transformation of adenosine (A) to m6A, whereas METTL14 is an essential component for supporting the recognition of RNA substrates by METTL3 (15-18). It is demonstrated that METTL3 is involved in the progression of liver cancer (19), breast cancer (20) and acute myeloid leukaemia (21). Recent studies have found that METTL14 also plays a critical role in acute myeloid leukaemia development and maintenance (15) as well as hepatocellular carcinoma metastasis (22). However, it is unclear whether the level of METTL14 impairs the growth and metastasis of cervical cancer.
Herein, we found that silencing METTL14 by shRNA-METTL14 interference inhibited the proliferation, migration and invasion of both HPV-positive and HPV-negative cervical cancer cells, and also triggered cell cycle arrest. This suggests a critical pro-oncogenic role of METTL14 in the progression of cervical cancer.
Methods
Cell culture and transfection
The human cervical cancer cell lines SiHa (HPV-positive) and C33a (HPV-negative) were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; HyClone, Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific) and antibiotics (Sigma-Aldrich, Germany) at 37 °C with 5% CO2. Once the cells entered the logarithmic growth phase, the pSuper-METTL14-shRNA (1# and 2#, 50 nM; Ribobio, China) vectors were constructed and transfected into SiHa and C33a cells using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific) to knock down the expression of METTL14, and the blank plasmid was used as the negative control (NC). This study is approved by the Medical Committee of Shandong Provincial Hospital Affiliated to Shandong University (No. 2019-146).
Real-time polymerase chain reaction (RT-PCR)
After 24 h of transfection, cells were harvested to extract the total RNA using Ultrapure RNA Kit (CWBIO, Beijing, China). Then, RNA was reverse transcribed to complementary cDNA using the HiFiScript cDNA Synthesis Kit (CWBIO, Beijing, China). The expression level of METTL14 was evaluated using the SYBR Premix Ex Taq II kit (Takara, Shiga, Japan) according to the manufacturer’s instructions. The primer sets were as follows: METTL14 F: 5'-GGGGTTGGACCTTGGAAGAG-3', R: 5'-TCCACCTCCTCGGTCAGATT-3'; β-actin F: 5'-CCCGAGCCGTGTTTCCT-3', R: 5'-GTCCCAGTTGGTGACGATGC-3'. Data were analyzed according to the sample threshold cycle (Ct) value from three independent experiments.
Western blot analysis
After 48 h of transfection, cells were collected and lysed in RIPA Lysis Buffer (CWBIO) for protein extraction and then quantified using a BCA kit (CWBIO). Equal amounts of protein (20 µg) were loaded and electrophoresed by 10% SDS-PAGE gel and electrotransferred to a polyvinylidene fluoride membrane (PVDF; Millipore, Billerica, MA, USA). The membrane was blocked with 5% non-fat milk for 1 h at room temperature followed by incubation with primary antibodies (dilution, 1:1,000; Proteintech Group, IL, USA) at 4 °C overnight. The membrane was washed with TBST (TBS buffer containing 0.01% Tween-20) buffer a total of three times, then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (dilution, 1:3,000; Proteintech Group) in blocking buffer for 1 h at room temperature. The membrane was visualized using the enhanced chemiluminescence kit (CWBIO), and GAPDH was used as an internal loading control.
Cell Counting Kit-8 (CCK8) assay
CCK8 (Beijing Solarbio Science & Technology, Beijing, China) was used to assess cell proliferation according to the manufacturer’s instructions. Cells transfected with sh-METTL14 for 24 h were seeded into a 96-well plate at a density of 1×103 cells per well. CCK8 reagent (10 µL) was added into each well and incubated at 37 °C for 1 h. Then, the OD value of excitation light was measured at 450 nm using an enzyme standard instrument. The assay was conducted three times independently.
Colony formation assay
Cells transfected with sh-METTL14 were seeded into plates at a density of 200 cells per well and cultured at 37 °C for about 1 week until visible colonies had formed. The colonies were fixed with 4% paraformaldehyde for 30 min, followed by staining with 0.1% crystal violet for 30 min. The assay was conducted three times independently.
Wound-healing assay
Following transfection with sh-METTL14 for 24 h, cells were cultured into 6-well plates at a density of 5×105/well for 12 h. A wound was formed with a pipette tip and cells were cultured in serum-free medium at 37 °C for 48 h. Wound healing was observed under a light microscope and quantitated with ImageJ software. The assay was independently conducted in triplicate.
Transwell invasion assay
Transwell chambers (Millipore, MA, USA) coated with Matrigel (BD Bioscience, CA, USA) were used for assessment of cell invasion according to the manufacturer’s instructions. Cells transfected with sh-METTL14 were collected and re-suspended in serum-free medium (1×106 cell/mL). A total of 100 µL of cell suspension (1×105 cells) was seeded into the upper compartment of the Transwell chambers and medium with 10% FBS was added into the lower chamber, which was followed by 24 h incubation. After incubation, non-invading cells were removed with a cotton-tipped swab, whereas invading cells were fixed with 4% paraformaldehyde for 30 min followed by staining with 0.1% crystal violet for 20 min. Images were captured of the stained cells (100× magnification) and cells were counted under a light microscope. The assay was conducted three times independently.
Cell cycle analysis
Flow cytometry was used to evaluate the cell cycle distribution. After 24 h of transfection with shRNA-METTL14, cells were fixed in pre-chilled 70% ethanol at −20 °C overnight, followed by staining with propidium iodide (PI) for 30 min at 37 °C. The cells were then analyzed by a flow cytometer (BD FACSC Anto II, BD Biosciences, USA). The assay was independently conducted in triplicate.
Statistical analysis
The data were expressed as the mean ± SD from the three independent experiments. All statistical data analyses were performed using GraphPad Prism 7.0 (GraphPad Soft-ware, Inc, La Jolla, CA, USA). The difference between two groups was estimated using the Student’s t-test. P<0.05 was considered statistically significant.
Results
Up-regulation of METTL14 is associated with a poor prognosis in patients with cervical cancer
Analysis from the Gene Expression Profiling Interactive Analysis (GEPIA) database (23) showed a positive correlation between the expression level of METTL14 mRNA and overall survival of patients with cervical cancer (P=0.04, Figure 1A). Furthermore, patients with high level of METTL14 showed a worse prognosis, indicating that METTL14 may play a role in the progression of cervical cancer.
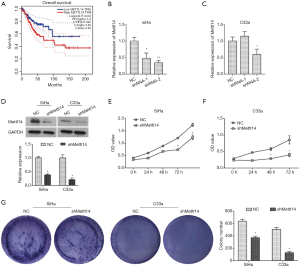
Silencing METTL14 in cervical cancer inhibits cell growth in vitro
In order to investigate the role of METTL14 in the cell growth of cervical cancer, two different shRNA sequences targeting METTL14 (shRNA-1# and shRNA-2#) were transfected into cervical cancer cell lines SiHa and C33a. This was performed to stably block the expression of METTL14. As shown in Figure 1B,C, transfection with shRNA-METTL14-2# significantly reduced the expression of METTL14 in both SiHa and C33a cells. Therefore, shRNA-METTL14-2# was used in all subsequent experiments due to its effective interference efficiency. Moreover, western blot analysis indicated that the expression of METTL14 was blocked by shMETTL14 in SiHa and C33a cells at the protein level (Figure 1D). CCK8 assay was used to assess the effect of silenced METTL14 on cell viability. As indicated in Figure 1E,F, compared to control groups, cell viability was significantly decreased in both shMETTL14-transfected SiHa and C33a cells (P<0.05). The inhibitory effect of silenced METTL14 on cervical cancer cell growth was further validated using colony formation assay. Compared with the control group, cell clonogenicity was significantly suppressed by shMETTL14 interference in SiHa and C33a cells (P<0.05, Figure 1G). Collectively, these data suggested that silencing METTL14 decreased cell growth of cervical cancer in vitro.
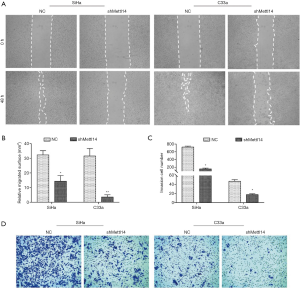
METTL14 knockdown suppresses cell migration and invasion abilities of cervical cancer in vitro
A wound-healing assay was carried out to determine whether METTL14 knockdown impacted cell migration in cervical cancer. As evident in Figure 2A,B, the migration ability of SiHa cells was significantly inhibited by shMETTL14 transfection compared to the control group (P<0.05), and a similar result was observed in C33a cells. In addition, METTL14-silenced SiHa and C33a cells both displayed a notable decrease in invasion compared to the control group (P<0.05, Figure 2C,D). These findings suggest that deficiency of METTL14 could result in a decrease in invasion ability of cervical cancer cells in vitro.
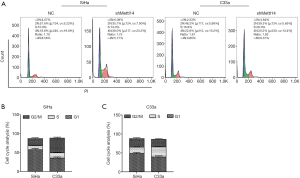
Silencing METTL14 triggers cell cycle arrest in cervical cancer cells
Flow cytometry was utilized to assess cell cycle distribution following METTL14 knock down. As shown in Figure 3A,B, the proportion of cells in the S and G2/M phases was increased in METTL14-knocked down SiHa cells compared to the NC group, and cells in G1 phase were decreased. A significant increase in the proportion of cells in the S phase was also observed in C33a cells transfected with shRNA-METTL14, but the proportion of cells that remained in G1 and G2/M phases decreased (Figure 3A,C). Our data indicated that METTL14 knockdown induced cell cycle arrest in cervical cancer cells, resulting in the inhibition of cell growth.
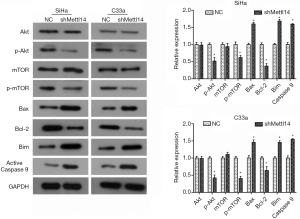
METTL14 knockdown down-regulates the PI3K/Akt/mTOR pathway in cervical cancer
To further investigate the mechanism underlying the suppression of cell growth and mobility caused by METTL14 deficiency in cervical cancer cells, changes in expression levels of PI3K/Akt/mTOR signaling pathway associated proteins were examined. As demonstrated by the results of western blot analysis, there was no significant change in the level of total Akt and mTOR after METTL14 was silenced in SiHa and C33a cells, but there was a significant decrease in the phosphorylation levels of Akt and Mtor (P<0.05, Figure 4). In addition, the expression level of its downstream apoptosis-related proteins also changed significantly. The expression levels of pro-apoptotic proteins Bax, Bim and active Caspase 9 were significantly up-regulated by METTL14 knockdown in SiHa and C33a cells, whereas the expression of anti-apoptotic protein Bcl-2 was down-regulated (P<0.05, Figure 4).
Discussion
Accumulating evidence has identified that RNA m6A modification is a type of epigenetic regulation which has shown potential for modulating mRNA decay, translation and processing by functional interactions between m6A methyltransferase and demethylase (22). Nevertheless, there is currently little research regarding the role of dysregulated m6A modification in human disease, especially in the occurrence and progression of cancer. METTL14, a methyltransferase which was first identified in mammals in 2014, can modify mammalian mRNA by catalyzing m6A methylation through forming a stable heterodimer with METTL3 (24,25). Here, we found that up-regulation of METTL14 acted as an adverse prognostic factor for overall survival in cervical cancer, indicating that dysregulation of METTL14 might be associated with the progression of cervical cancer. Therefore, METTL14 was knocked down by shRNA-METTL14 interference in cervical cancer cell lines SiHa and C33a, and further studies were performed to assess the effect of METTL14 knockdown on cellular behaviors of SiHa and C33a cells. In the current study, our data revealed that METTL14 knockdown suppressed the proliferation, migration and invasion abilities of both SiHa and C33a cells in vitro, suggesting that METTL14 might function as an oncogene in cell growth and metastasis of HPV-positive and HPV-negative cervical cancer.
The level of m6A varies in different tissues and cell lines, with increased expression in the liver, kidney and brain (22,26). Weng et al. reported that METTL14 is up-regulated in human acute myeloid leukemia cell lines, and METTL14 knockdown could repress cell survival and proliferation and promote terminal myeloid differentiation (15). It has also been revealed that down-regulation of m6A by METTL3 and METTL14 knockdown could suppress cell viability in HeLa cells (24). However, decreased tendency of m6A was observed in hepatocellular carcinoma, which is mainly due to the down-regulation of METTL14, and up-regulation of METTL14 inhibits tumor metastasis and silencing of METTL14 increases tumor metastasis (22). Therefore, the specific levels of m6A appear to play different roles in different tissues and cell lines. In our study, we confirmed that METTL14 knockdown repressed growth and invasion of cervical cancer cells in vitro, which was consistent with the findings in acute myeloid leukemia and Hela cells.
It is well known that cell cycle dysregulation is one of the most common features of cancer cells, including cervical cancer. Considering the effect of METTL14 knockdown on the proliferation of cervical cancer cells, we assessed the effect of METTL14 silencing on cell cycle distribution. Here, for the first time, our data showed that METTL14 knockdown induced cell cycle arrest during S phase in SiHa and C33a cells by promoting more cells to stay in S phase, resulting in decreased cells in the G1 and G2/M phases in C33a cells, but increased the proportion of cells in the G2/M phase in SiHa cells. The difference in cell cycle distribution caused by METTL14 knockdown could be due to the diversity of cell lines, which requires further study.
In the current study, our data revealed an oncogenic role of METTL14 in the growth and invasion of cervical cancer. Therefore, we sought to investigate the mechanism of METTL14 underlying the regulation of cervical cancer progression. The PI3K/Akt/mTOR signaling pathway plays an essential role in many cellular processes as well as tumorigenesis and development, such as cell survival and proliferation, cell cycle regulation and invasion (27). Therefore, blocking this pathway is an important method for suppressing tumor growth. Thus, we evaluated whether the expression levels of key components of the PI3K/Akt/mTOR signaling pathway changed after METTL14 silencing. As indicated by western blot analysis, METTL14 knockdown suppressed the PI3K/Akt/mTOR pathway by down-regulating the phosphorylation levels of Akt and mTOR. In addition, the expression of apoptosis-related proteins was also affected in METTL14 silenced cells, indicating that loss of METTL14 promoted cell apoptosis by regulating the Bcl-2/Bax axis and the Caspase cascade. Collectively, the PI3K/Akt/mTOR signaling pathway might be involved in the regulation of METTL14 on the growth and invasion of cervical cancer cells. Recent studies have confirmed that the functional mechanism of METTL14 is dependent on regulation through m6A modification (15,22). Weng et al. revealed that METTL14 modulates myeloid differentiation and self-renewal and growth of normal HSPCs or LSCs/LICs cells by regulating MYB and MYC through m6A modification (15). Ma et al. show that METTL14 modulates the primary miRNA126 process in an m6A-dependent manner in SMMC-7721, HepG2 and HCC-1664 cells (22). Based on these results, further studies are necessary to identify the target genes of METTL14 in order to investigate the molecular mechanism of its oncogenic role in cervical cancer.
Conclusions
In summary, the current study demonstrates the oncogenic role of METTL14 in HPV-positive and HPV-negative cervical cancer cells. Targeting METTL14 could suppress cell proliferation, migration and invasion in SiHa and C33a cells, as well as trigger cell cycle arrest. Moreover, silencing METTL14 also inhibits the PI3K/Akt/mTOR signaling pathway, which might be involved in the function of METTL14. Our study suggests a novel potential target for the treatment of HPV-positive and HPV-negative cervical cancer.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.48). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study is approved by the Medical Committee of Shandong Provincial Hospital Affiliated to Shandong University (No. 2019-146). Individual informed consent was waived due to the nature of the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Small W Jr, Bacon MA, Bajaj A, et al. Cervical cancer: A global health crisis. Cancer 2017;123:2404-12. [Crossref] [PubMed]
- Yang M, Zhai X, Ge T, et al. MiR-181a-5p Promotes Proliferation and Invasion, and Inhibits Apoptosis of Cervical Cancer Cells via Regulating Inositol Polyphosphate-5-Phosphatase A (INPP5A). Oncol Res 2018;26:703-12. [Crossref] [PubMed]
- Pötter R, Georg P, Dimopoulos JC, et al. Clinical outcome of protocol based image (MRI) guided adaptive brachytherapy combined with 3D conformal radiotherapy with or without chemotherapy in patients with locally advanced cervical cancer. Radiother Oncol 2011;100:116-23. [Crossref] [PubMed]
- Sturdza A, Pötter R, Fokdal LU, et al. Image guided brachytherapy in locally advanced cervical cancer: Improved pelvic control and survival in RetroEMBRACE, a multicenter cohort study. Radiother Oncol 2016;120:428-33. [Crossref] [PubMed]
- Sankaranarayanan R, Swaminathan R, Brenner H, et al. Cancer survival in Africa, Asia, and Central America: a population-based study. Lancet Oncol 2010;11:165-73. [Crossref] [PubMed]
- Gershenson DM, Mcguire WP, Gore M, et al. Gynecologic Cancer: Controversies in Management. 1st edition. London: Churchill Livingstone, 2004.
- Crosbie EJ, Kitchener HC. Human papillomavirus in cervical screening and vaccination. Clin Sci (Lond) 2006;110:543-52. [Crossref] [PubMed]
- de Sanjose S, Quint WG, Alemany L, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol 2010;11:1048-56. [Crossref] [PubMed]
- Wang HY, Kim G, Cho H, et al. Diagnostic performance of HPV E6/E7, hTERT, and Ki67 mRNA RT-qPCR assays on formalin-fixed paraffin-embedded cervical tissue specimens from women with cervical cancer. Exp Mol Pathol 2015;98:510-6. [Crossref] [PubMed]
- Zhao X, Yang Y, Sun BF, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res 2014;24:1403-19. [Crossref] [PubMed]
- Deng X, Su R, Feng X, et al. Role of N6-methyladenosine modification in cancer. Curr Opin Genet Dev 2018;48:1-7. [Crossref] [PubMed]
- Wang X, Zhao BS, Roundtree IA, et al. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015;161:1388-99. [Crossref] [PubMed]
- Batista PJ, Molinie B, Wang J, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014;15:707-19. [Crossref] [PubMed]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 2015;347:1002-6. [Crossref] [PubMed]
- Weng H, Huang H, Wu H, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A Modification. Cell Stem Cell 2018;22:191-205.e9. [Crossref] [PubMed]
- Śledź P, Jinek M. Structural insights into the molecular mechanism of the m6A writer complex. Elife 2016; [Crossref] [PubMed]
- Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell 2016;63:306-17. [Crossref] [PubMed]
- Wang X, Feng J, Yuan X, et al. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 2017;542:260. [Crossref] [PubMed]
- Chen M, Wei L, Law CT, et al. RNA N6‐methyladenosine methyltransferase‐like 3 promotes liver cancer progression through YTHDF2‐dependent posttranscriptional silencing of SOCS2. Hepatology 2018;67:2254-70. [Crossref] [PubMed]
- Cai X, Wang X, Cao C, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett 2018;415:11-9. [Crossref] [PubMed]
- Barbieri I, Tzelepis K, Pandolfini L, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 2017;552:126-31. [Crossref] [PubMed]
- Ma JZ, Yang F, Zhou CC, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6 -methyladenosine-dependent primary MicroRNA processing. Hepatology 2017;65:529-43. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 2014;10:93-5. [Crossref] [PubMed]
- Wang Y, Li Y, Toth JI, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol 2014;16:191-8. [Crossref] [PubMed]
- Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 2012;149:1635-46. [Crossref] [PubMed]
- Kwong LN, Davies MA. Navigating the therapeutic complexity of PI3K pathway inhibition in melanoma. Clin Cancer Res 2013;19:5310-9. [Crossref] [PubMed]


