
LncRNA-DANCR promotes growth and metastasis of colorectal cancer via activating epithelial-mesenchymal transition process
Introduction
Colorectal cancer (CRC) is one of the most common malignant cancers in the world, and its annual fatality is the fourth highest in the world (1-3). Although the diagnosis and treatment strategies have improved in recent years, the cure rate remains at a low level. The main reason is that tumor metastasis dramatically increases the recurrence rate (4). So far, the underlying mechanism of CRC metastasis is not clear. Therefore, it is urgent to investigate novel biomarkers and mechanisms for the targeted therapy.
Long non-coding RNAs (lncRNAs) are identified as endogenous cellular RNAs with more than 200 nucleotides in length and lack an open reading frame (5-7). In recent years, more and more studies have shown that lncRNAs play critical roles in many physiological and pathological processes including immune regulation, embryonic development and tumorigenesis (8,9). Zhou et al. (10) have reported that lncRNA-GACAT3 could promote cell proliferation and migration in CRC. LncRNA SLCO4A1-AS1 facilitates the growth and metastasis of CRC through β-catenin-dependent Wnt pathway (11). Therefore, the functions of lncRNA in the growth and metastasis of CRC have become a popular research direction. Long non-coding RNA-differentiation antagonizing non-protein coding RNA (DANCR) is a promising tumor-associated lncRNA, which is up-regulated in various human cancers (12). Liu et al. (13) found that the high expression of lncRNA-DANCR was involved in the progression of CRC. In addition, it has been confirmed that lncRNA-DANCR could promote cell proliferation and metastasis via miR-577 sponging in CRC (14). However, the exact mechanism of lncRNA-DANCR in CRC remains unclear.
The activation of epithelial-mesenchymal transition (EMT) plays a key role in metastatic cascade by increasing cell migration and invasion. EMT is characterized by up-regulation of mesenchymal markers such as vimentin and down-regulation of epithelial markers such as E-cadherin (15,16). In addition, recent studies have confirmed that EMT is a key factor in the distant metastasis of CRC (17). Several researches have confirmed that lncRNAs mediated the growth and metastasis of tumor by regulating EMT process of tumor cells. LncRNA aquaporin could promote the migration and metastasis of intestinal cancer cells via EMT process (18). Guo et al. (19) have found in their research that lncRNA-DANCR could promote the migration of lung cancer cells by activating the EMT process. Therefore, we investigated whether lncRNA-DANCR promotes the growth and metastasis of CRC by activating the EMT process.
Methods
Human tissue specimen
A total of 40 CRC tissues and paired adjacent non-cancerous tissue samples were obtained from patients who underwent surgical resection of CRC in our hospital from 2016 to 2018. All tissue samples were collected and stored at −80 °C. This study was approved by the Ethics Committee of our hospital and was conducted in accordance with the Helsinki Declaration and the guidelines of the International Rehabilitation Council for Torture Victims. All the patients recruited signed informed consent.
Cell cultures and transfection
Human CRC cell HT-29 and normal colorectal epithelial cell FHC were cryopreserved in our laboratory. All the cells were cultivated in Dulbecco’s modified Eagle’s medium (DMEM, Sigma, USA) containing 10% fetal bovine serum (FBS, Gibco, USA), at 37 °C in 5% carbon dioxide and 95% air.
The si-DANCR interference sequences and the si-NC sequences (empty sequences without the interference) were transfected into HT-29 cells by Lipofectamine® 2000 Reagent (Invitrogen, USA). In short, HT-29 cells were added to 6-well plates and then transfected with 0.1 ng siRNA or si-NC when cells were 80% confluence in the plate well. The transfected HT-29 cells were randomly divided into three groups: BLANK group (cultured with medium only), si-NC group (cultured with si-NC sequence) and si-DANCR group (cultured with si-DANCR interference sequence). The primer sequences were designed and synthesized by Invitrogen (Shanghai, China) as shown in Table 1. Finally, all the cells were cultured in 37 °C incubator for 48 h.
Table 1
| Name of primer | Sequences (5'-3') |
|---|---|
| si-DANCR-F | CGUACUAACUUGUAGCAACCA |
| si-DANCR-R | GUUGCUACAAGUUAGUACGCA |
| si-NC-F | UUCUCCGAACGUGUCACGUTT |
| si-NC-R | ACGUGACACGUUCGGAGAATT |
MTT assays
Cell viability was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma, USA) assay. Three groups of HT-29 cells were inoculated into 96 well plates (3,000–6,000 cells/well). At different time points (days 0, 1, 2 and 3), the culture medium was removed and 20 µl of MTT (5 mg/mL) was added into each well. After incubation at 37 °C for 4 h, MTT was removed, and absorbance at 490 nm were measured on the microplate reader (Bio-Rad, USA).
Wound healing assays
Wound healing assays were performed as previously described by (20,21). HT-29 cells were inoculated onto 6-well plates (5×105 cells/well) and cultivated at 37 °C. After 24 h, the wound was scratched with a 20 µL pipette tip and washed three times with PBS. The cells were then cultured in Serum free medium at 37 °C. Subsequently, the wound width was then measured under light microscopy at 0 and 24 h.
Transwell assays
Transwell assay was performed as previously described (22,23). HT-29 cells (5×105 cells/well) were inoculated to an upper chamber (Corning, USA) and the inserts were coated with Matrigel (BD Biosciences, USA). Six hundred µL of complete medium was present in that bottom chamber. After incubation in 37 °C incubator for 24 h, the non-migratory cells were removed in the upper chamber. The migrated cells were fixed with 4% paraformaldehyde and then stained with crystal violet. The migrating cells were counted in 5 random fields under an optical microscope.
Real-time fluorogenic PCR assays
Total RNA was extracted using TRIZOL (Invitrogen, USA), reverse-transcribed into cDNA by Revert Aid First Strand cDNA Synthesis Kit (ThermoScientific, USA) and measured by using qRT-PCR (Bio-Rad, USA) with SYBR green qPCR Master Mix (Thermo Scientific, USA). Primers used for qRT-PCR analysis were the following: DANCR (F: 5'-GCGCCACTATGTAGCGGGTT-3', R: 5'-TCAATGGCTTGTGCCTGTAGTT-3'); GAPDH (F: 5'-GTCGATGGCTAGTCGTAGCATCGAT-3', R: 5'- TGCTAGCTGGCATGCCCGATCGATC-3').
Western blot analysis
Total proteins were extracted by lysis buffer. Protein samples (30 µg) were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene difluoride (PVDF) membrane. The membranes were sealed with 5% skimmed milk and incubated overnight with primary antibodies (E-cadherin, 1:1,000; Vimentin, 1:1,000; GAPDH, 1:3,000, Cell Signal, USA) at 4 °C. Afterwards, the peroxidase-labeled secondary antibody (anti-rabbit IgG, 1:5,000, Cell Signal, USA) was used for incubation for 2 h. The protein blots were visualized with an enhanced chemiluminescence (ECL) kit. Finally, the density of western blot bands was analyzed using Quantity One 1-D Analysis Software (Bio-Rad, USA).
Immunofluorescence analysis
HT-29 cells were collected and inoculated onto the 12-well plates (5×104 cells/well). After 24 h, the cells were washed with PBS three times and 4% paraformaldehyde was added to fix the cells. After 0.4% Triton X-100 was added, the cells were blocked by adding 5% BSA for 30 min, and then incubated overnight at 4 °C with the primary antibodies (E-cadherin, 1:100, Santa Cruz, China; Vimentin, 1:100, Cell Signal, USA). Afterwards, the cells were incubated with Alexa Fluor 488-conjugated secondary antibody (1:500, Invitrogen, USA) in a dark room for 1 h and then washed three times with PBS. Finally, the cells were stained with 4',6-diamidino-2-phenylindole (DAPI, Sigma, Germany) for 5 min and analyzed by the Carl Zeiss Confocal Imaging System (LSM 780, Carl Zeiss, Germany).
Mice model establishment and hematoxylin-eosin (HE) staining
Male nude mice (20–25 g, 4–6 weeks) were provided by the Laboratory Animal Center of our hospital. All animal assays were approved by the Ethics Committee of our hospital. The xenograft tumor mice model was established by intravenous injection of transfected HT-29 cells (1×106 cells, 100 µL). The mice were randomly divided into three groups (n=6 per group): the BLACK group (injected cells cultured with medium only), si-NC group (injected cells transfected with si-NC sequence) and si-DANCR group (injected cells transfected with si-DANCR sequence). Seven weeks after injection, the animals were euthanized and lung lobes were removed. The lung lobes were embedded in paraffin, sectioned, and stained with HE. Visible lung metastatic nodules were then observed under a light microscope.
Statistical analysis
All statistical analyses were performed using SPSS17.0 Statistical Software (Chicago, IL). The results were presented in the form of mean ± standard deviation. The differences between various groups were analyzed by one-way ANOVA followed by the Tukey’s post hoc test, and the data of the two groups was assessed using the Student’s t-test. P<0.05 was considered to be statistically significant.
Results
The expression of lncRNA-DANCR in human CRC tissues and HT-29 cell lines
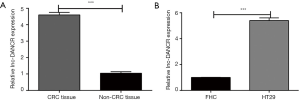
As shown in Figure 1A, the lncRNA-DANCR expression apparently increased in human CRC tissues in comparison with non-CRC tissues (P<0.001). Figure 1B showed that the mRNA expression of lncRNA-DANCR in HT-29 cells was also significantly higher than that in FHC cells (P<0.001).
Cell transfection efficiency
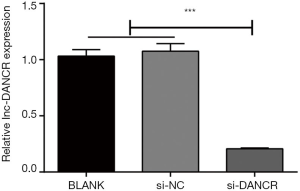
After 48 hours of transfection, lncRNA-DANCR expressions significantly decreased in si-DANCR group compared with BLANK and si-NC group (P<0.001, Figure 2). All those results revealed si-DANCR interference sequence were successfully transfected into HT-29 cells.
LncRNA-DANCR increased the viability and migration of HT-29 cells
As shown in Figure 3A, compared with BLANK and si-NC group, the proliferation ability of si-DANCR group was significantly inhibited at 1 d (P<0.01), 2 d (P<0.001) and 3 d (P<0.001). Wound healing assays results showed the migration ability was markedly inhibited in si-DANCR group compared to BLANK and si-NC group (P<0.001, Figure 3B,C).

LncRNA-DANCR increased HT-29 cells invasion ability
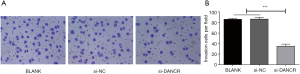
After 24 hours of transfection, the numbers of HT-29 cells invasions in BLANK, si-NC and si-DANCR group were 85±5, 87±5 and 37±4, respectively. The invasion ability was dramatically inhibited in si-DANCR group compared to BLANK and si-NC group (P<0.001, Figure 4A,B).
LncRNA-DANCR induced EMT process in HT-29 cells
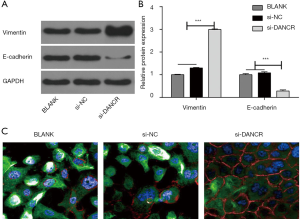
After 72 hours of transfection, the expressions of EMT-related proteins (E-cadherin and Vimentin) were measured by Western blot analysis and immunofluorescence analysis. As seen in Figure 5A,B, the protein expression of vimentin in si-DANCR group was inhibited dramatically, compared with BLANK and si-NC group (P<0.001). On the contrary, E-cadherin protein expression was significantly increased in si-DANCR group compared with BLANK and si-NC group (P<0.001). The same results were observed in the immunofluorescence experiment (Figure 5C).
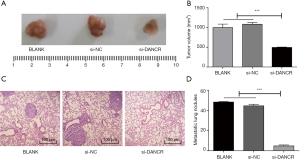
LncRNA-DANCR promoted CRC metastasis in vivo
To further study whether lncRNA-DANCR can affect the metastasis process in vivo, a mouse model of xenograft tumor was established. As shown in Figure 6A,B, silencing DANCR significantly reduced the tumor volume in nude mice. HE staining results showed that silencing DANCR significantly decreased metastatic lung nodules compared with BLANK and si-NC group (P<0.001) (Figure 6C,D).
Discussion
CRC is one of the most common cancers around the world (24). Approximately 1.4 million new cases are identified and almost 694,000 deaths are reported each year (25). Therefore, it is urgent to explore and find new molecular approaches to better understand this disease and identify new therapeutic targets. This study suggests that lncRNA-DANCR may promote the growth and metastasis of CRC by activating the EMT process.
More and more evidences show that lncRNAs are involved in many biological processes and play an important role in the development of diseases (26,27). Abnormal expression of lncRNAs might directly influence the onset and progression of tumors. Recent studies have shown that a variety of lncRNAs play an important role in CRC, thus revealing their potential as novel therapeutic targets. Many researches have suggested that various lncRNAs such as lncRNA SNHG6, lncRNA LOC101927746 and LncRNA SLCO4A1-AS1 are abnormally expressed in CRC (11,28,29). Liu et al. (13) have reported that lncRNA-DANCR is highly expressed in CRC. Our results also confirmed that in human CRC tissues and HT-29 cells, lncRNA-DANCR expression was apparently higher than that in non-CRC tissues and FHC cells. However, it is unclear how lncRNA-DANCR affects the development of CRC. Liu et al. (30) have indicated that lncRNA DLEU1 protects the CRC progression by regulating KPNA3. Gu et al. (31) have reported that long noncoding RNA BCYRN1 promotes the CRC growth by inhibiting NPR3. Lan et al. (32) have proved that the lncRNA OCC-1 inbibits CRC cells proliferation by destabilizing HuR protein. Yu et al. (11) have confirmed that lncRNA SLCO4A1-AS1 promotes the growth of CRC through regulating Wnt/β-catenin pathway. These studies above have suggested that intensive studies of lncRNAs functions are helpful for the diagnostic and therapeutic strategies of CRC. It is reported that lncRNA-DANCR may facilitate CRC cells proliferation and metastasis in CRC (14). Our results showed that silencing lncRNA-DANCR significantly inhibited HT-29 cells proliferation at 1 d, 2 d and 3 d. Moreover, silencing lncRNA-DANCR markedly also suppressed HT-29 cells invasion and metastasis. It was confirmed that lncRNA-DANCR significantly promoted the growth and metastasis of CRC. Interestingly, lung metastasis, being considered as the common target organ, was observed in xenograft tumor mice model. Our study suggests that lncRNA-DANCR could promote lung metastasis and lead to poor disease prognosis.
But how does lncRNA-DANCR exert its functions? That needs to be further explored. It has been preliminarily confirmed by researchers that EMT plays an important role in tumor metastatic by regulating cell migration and invasion D C BramB Geert (33). EMT is a process in which tumor cells differentiate into mesenchymal cells. Tumor cells undergoing EMT process exhibits both morphological and molecular changes, as demonstrated by the low expression of epithelial markers (E-cadherin), and the high expression of mesenchymal markers (Vimentin). Gao et al. (34) have confirmed that lncRNAH19 mediates the chemo-sensitivity of breast cancer cells via the EMT process. Li et al. indicate that long noncoding RNA XLOC 010588 could regulate the progression of CRC via the EMT process (35). It is reported that lncRNA PlncRNA-1 could induce the activation of epithelial mesenchymal transition in CRC cells (36). Moreover, Guo et al. (19) have found that long noncoding RNA DANCR could promote the progression of lung cancer by activating the EMT process. Therefore, we investigated whether lncRNA-DANCR might promote the growth and metastasis of CRC by activating the EMT process. Both western blot and immunofluorescence analysis results showed that silencing lncRNA-DANCR significantly increased the expression of E-cadherin. On the contrary, silencing lncRNA-DANCR markedly inhibited vimentin expression. Those evidence suggested that lncRNA-DANCR could promote the growth and metastasis of by activating the EMT process.
Conclusions
We demonstrated that lncRNA-DANCR could facilitate the growth and metastasis of CRC by regulating the EMT process, suggesting that lncRNA-DANCR may serve as a bio-marker or target for CRC metastasis.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.10.09). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of our hospital and was conducted in accordance with the Helsinki Declaration (as revised in 2013) and the guidelines of the International Rehabilitation Council for Torture Victims. All the patients recruited signed informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Arnold M, Sierra M S, Laversanne M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017;66:683-91. [Crossref] [PubMed]
- Chen X, Liu B, Yang R, et al. Integrated analysis of long non-coding RNAs in human colorectal cancer. Oncotarget 2016;7:23897-908. [PubMed]
- Wang F, Ni H, Sun F, et al. Overexpression of lncRNA AFAP1-AS1 correlates with poor prognosis and promotes tumorigenesis in colorectal cancer. Biomed Pharmacother 2016;81:152-9. [Crossref] [PubMed]
- Compton CC. Colorectal Carcinoma: Diagnostic, Prognostic, and Molecular Features. Mod Pathol 2003;16:376-88. [Crossref] [PubMed]
- Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007;316:1484-8. [Crossref] [PubMed]
- Ponting CP, Oliver P. LReik W. Evolution and Functions of Long Noncoding RNAs. Cell 2009;136:629-41. [Crossref] [PubMed]
- Guttman M, Garber M, Levin JZ, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 2010;28:503-10. [Crossref] [PubMed]
- Karlic R, Ganesh S, Franke V, et al. Long non-coding RNA exchange during the oocyte-to-embryo transition in mice. DNA Res 2017;24:129-41. [Crossref] [PubMed]
- Liu B, Ye B, Yang L, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol 2017;18:499-508. [Crossref] [PubMed]
- Zhou W, Wang L, Miao Y, et al. Novel long noncoding RNA GACAT3 promotes colorectal cancer cell proliferation, invasion, and migration through miR-149. Onco Targets Ther 2018;11:1543-52. [Crossref] [PubMed]
- Yu J, Han Z, Sun Z, et al. LncRNA SLCO4A1-AS1 facilitates growth and metastasis of colorectal cancer through β-catenin-dependent Wnt pathway. J Exp Clin Cancer Res 2018;37:222. [Crossref] [PubMed]
- Thin KZ, Liu X, Feng X, et al. LncRNA-DANCR: A valuable cancer related long non-coding RNA for human cancers. Pathol Res Pract 2018;214:801-5. [Crossref] [PubMed]
- Liu Y, Zhang M, Liang L, et al. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol 2015;8:11480-4. [PubMed]
- Wang Y, Lu Z, Wang N, et al. Long noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 sponging. Exp Mol Med 2018;50:57. [Crossref] [PubMed]
- Garg M. Epithelial-mesenchymal transition-activating transcription factors-multifunctional regulators in cancer. World J Stem Cells 2013;5:188-95. [Crossref] [PubMed]
- Hu Y, Dai M, Zheng Y, et al. Epigenetic suppression of E-cadherin expression by Snail2 during the metastasis of colorectal cancer. Clin Epigenetics 2018;10:154. [Crossref] [PubMed]
- Loboda A, Nebozhyn MV, Watters JW, et al. EMT is the dominant program in human colon cancer. BMC Med Genomics 2011;4:9. [Crossref] [PubMed]
- Chen J, Wang T, Zhou YC, et al. Aquaporin 3 promotes epithelial-mesenchymal transition in gastric cancer. J Exp Clin Cancer Res 2014;33:38. [Crossref] [PubMed]
- Guo L, Gu J, Hou S, et al. Long non-coding RNA DANCR promotes the progression of non-small-cell lung cancer by inhibiting p21 expression. Onco Targets Ther 2018;12:135-46. [Crossref] [PubMed]
- Shimada H, Abe S, Kohno T, et al. Loss of tricellular tight junction protein LSR promotes cell invasion and migration via upregulation of TEAD1/AREG in human endometrial cancer. Sci Rep 2017;7:37049. [Crossref] [PubMed]
- Lu D, Yao Q, Zhan C, et al. MicroRNA-146a promote cell migration and invasion in human colorectal cancer via carboxypeptidase M/src-FAK pathway. Oncotarget 2017;8:22674-84. [PubMed]
- Tiwari A, Pattnaik N, Jaiswal AM, et al. Increased FSHD region gene1 expression reduces in vitro cell migration, invasion, and angiogenesis, ex vivo supported by reduced expression in tumors. Biosci Rep 2017; [Crossref] [PubMed]
- Verma S, Pal R, Gupta SK. Decrease in invasion of HTR-8/SVneo trophoblastic cells by interferon gamma involves cross-communication of STAT1 and BATF2 that regulates the expression of JUN. Cell Adh Migr 2018;12:432-46. [PubMed]
- Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin 2014;64:104-17. [Crossref] [PubMed]
- Torre LA, Freddie B, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Wu H, Yang L, Chen LL. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet 2017;33:540-52. [Crossref] [PubMed]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013;152:1298-307. [Crossref] [PubMed]
- Xu M, Chen X, Lin K, et al. lncRNA SNHG6 regulates EZH2 expression by sponging miR-26a/b and miR-214 in colorectal cancer. J Hematol Oncol 2019;12:3. [Crossref] [PubMed]
- Huang H, Cai L, Li R, et al. A novel lncRNA LOC101927746 accelerates progression of colorectal cancer via inhibiting miR-584-3p and activating SSRP1. Biochem Biophys Res Commun 2019;509:734-8. [Crossref] [PubMed]
- Liu T, Han Z, Li H, et al. LncRNA DLEU1 contributes to colorectal cancer progression via activation of KPNA3. Mol Cancer 2018;17:118. [Crossref] [PubMed]
- Gu L, Lu L, Zhou D, et al. Long Noncoding RNA BCYRN1 Promotes the Proliferation of Colorectal Cancer Cells via Up-Regulating NPR3 Expression. Cell Physiol Biochem 2018;48:2337-49. [Crossref] [PubMed]
- Lan Y, Xiao X, He Z, et al. Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res 2018;46:5809-21. [Crossref] [PubMed]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013;13:97-110. [Crossref] [PubMed]
- Gao H, Hao G, Sun Y, et al. Long noncoding RNA H19 mediated the chemosensitivity of breast cancer cells via Wnt pathway and EMT process. Onco Targets Ther 2018;11:8001-12. [Crossref] [PubMed]
- Li Y, Zhao L, Zhang Y, et al. Downregulation of the long non-coding RNA XLOC_010588 inhibits the invasion and migration of colorectal cancer. Oncol Rep 2018;39:1619-30. [PubMed]
- Jia GQ, Zhang MM, Wang K, et al. Long non-coding RNA PlncRNA-1 promotes cell proliferation and hepatic metastasis in colorectal cancer. J Cell Biochem 2018;119:7091-104. [Crossref] [PubMed]


